

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.18 - phase: 2 /pseudo
(374 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
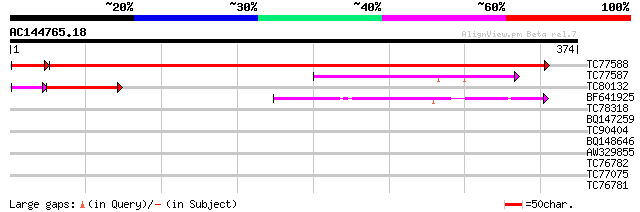
Score E
Sequences producing significant alignments: (bits) Value
TC77588 similar to PIR|D96538|D96538 cytosolic tRNA-Ala syntheta... 643 0.0
TC77587 weakly similar to PIR|D96538|D96538 cytosolic tRNA-Ala s... 117 9e-27
TC80132 similar to PIR|D96538|D96538 cytosolic tRNA-Ala syntheta... 91 4e-20
BF641925 similar to GP|10177227|db alanyl-tRNA synthetase {Arabi... 76 2e-14
TC78318 similar to PIR|T45852|T45852 hypothetical protein F3A4.7... 34 0.10
BQ147259 similar to PIR|C96654|C96 hypothetical protein F16P17.1... 29 3.4
TC90404 similar to GP|21593619|gb|AAM65586.1 receptor protein ki... 28 4.4
BQ148646 weakly similar to GP|11596186|gb| cystatin-like protein... 28 5.7
AW329855 similar to PIR|T51383|T51 receptor protein kinase-like ... 28 5.7
TC76782 similar to GP|12539609|gb|AAG59584.1 pathogen-inducible ... 28 5.7
TC77075 similar to GP|21593294|gb|AAM65243.1 putative carboxyl-t... 28 5.7
TC76781 similar to GP|12539609|gb|AAG59584.1 pathogen-inducible ... 28 5.7
>TC77588 similar to PIR|D96538|D96538 cytosolic tRNA-Ala synthetase
[imported] - Arabidopsis thaliana, partial (64%)
Length = 2100
Score = 643 bits (1658), Expect(2) = 0.0
Identities = 325/330 (98%), Positives = 326/330 (98%)
Frame = +2
Query: 27 NNDDPTCIEIWNLVLXQFNREADSSLKSLPAKHVDTGMGFERLTSILQRKLSNYDTDVFM 86
NNDDPTCIEIWNLV QFNREADSSLKSLPAKHVDTGMGFERLTSILQRKLSNYDTDVFM
Sbjct: 131 NNDDPTCIEIWNLVFIQFNREADSSLKSLPAKHVDTGMGFERLTSILQRKLSNYDTDVFM 310
Query: 87 PIFDAIQLATGARPYSGKVGPEDADKIDMAYRVVADHIRTLSFAIADGSRPGNEGREYVL 146
PIFDAIQLATGARPYSGKVGPEDADKIDMAYRVVADHIRTLSFAIADGSRPGNEGREYVL
Sbjct: 311 PIFDAIQLATGARPYSGKVGPEDADKIDMAYRVVADHIRTLSFAIADGSRPGNEGREYVL 490
Query: 147 RRILRRAVRYGREVLKAEEGFFNGLVNVVANVLGDVFPELKQQEVHIRNVIQEEEESFGR 206
RRILRRAVRYGREVLKAEEGFFNGLVNVVANVLGDVFPELKQQEVHIRNVIQEEEESFGR
Sbjct: 491 RRILRRAVRYGREVLKAEEGFFNGLVNVVANVLGDVFPELKQQEVHIRNVIQEEEESFGR 670
Query: 207 TLVKGIAKFKTAVGHVQGNILSGEATFELWDTYGFPLDLTQLMAEEKKLKVDVEGFNSAM 266
TLVKGIAKFKTAVGHVQGNILSGE FELWDTYGFPLDLTQLMAEEKKLKVDVEGFNSAM
Sbjct: 671 TLVKGIAKFKTAVGHVQGNILSGEVAFELWDTYGFPLDLTQLMAEEKKLKVDVEGFNSAM 850
Query: 267 EAARERSRTAQNKQAGGAIVMDADATSALQKRSIVPTDDSFKFASFRDHETVVKAIYTGS 326
EAARERSRTAQNKQAGGAIVMDADATSALQKRSIVPTDDSFKFASFRDHETVVKAIYTGS
Sbjct: 851 EAARERSRTAQNKQAGGAIVMDADATSALQKRSIVPTDDSFKFASFRDHETVVKAIYTGS 1030
Query: 327 EFVDSVNTDGDVGVILESTSFYAEQGGQVF 356
EFVDSVNTDGDVGVILESTSFYAEQGGQ+F
Sbjct: 1031EFVDSVNTDGDVGVILESTSFYAEQGGQIF 1120
Score = 51.6 bits (122), Expect(2) = 0.0
Identities = 21/25 (84%), Positives = 21/25 (84%)
Frame = +1
Query: 2 *HPSXWTLHRNSFXQXWQPGCCIFG 26
*H S WTLHRNSF Q WQPGCCIFG
Sbjct: 55 *HRSLWTLHRNSF*QNWQPGCCIFG 129
>TC77587 weakly similar to PIR|D96538|D96538 cytosolic tRNA-Ala synthetase
[imported] - Arabidopsis thaliana, partial (21%)
Length = 1024
Score = 117 bits (292), Expect = 9e-27
Identities = 69/142 (48%), Positives = 83/142 (57%), Gaps = 6/142 (4%)
Frame = +1
Query: 201 EESFGRTLVKGIAKFKTAVGHVQGNILSGEATFELWDTYGFPLDLTQLMAEEKKLKVDVE 260
EESF R L KGI KF AV HVQGNILSGE FEL T+GFPLDLTQLMAEEKK VD+E
Sbjct: 1 EESFSRNLFKGIRKFNKAVEHVQGNILSGEVAFELSHTFGFPLDLTQLMAEEKKFSVDIE 180
Query: 261 GFNSAMEAARERSRTAQNKQA----GGAIVMDADATSALQKR--SIVPTDDSFKFASFRD 314
GF+ AM+AARERS+TA K+ + A + + ++P D K
Sbjct: 181 GFHRAMKAARERSKTAPKKKVLSLKSNVETLSIPAAKKAENKIEIVLPQDQMRKAQKHVA 360
Query: 315 HETVVKAIYTGSEFVDSVNTDG 336
E KA+ +E D +DG
Sbjct: 361 EEDKRKAVKITTEKADLAVSDG 426
>TC80132 similar to PIR|D96538|D96538 cytosolic tRNA-Ala synthetase
[imported] - Arabidopsis thaliana, partial (25%)
Length = 908
Score = 91.3 bits (225), Expect(2) = 4e-20
Identities = 42/50 (84%), Positives = 45/50 (90%)
Frame = +1
Query: 25 FGNNDDPTCIEIWNLVLXQFNREADSSLKSLPAKHVDTGMGFERLTSILQ 74
F N++DP CIE+WNLV QFNREAD SLKSLPAKHVDTGMGFERLTSILQ
Sbjct: 757 FVNDEDPNCIELWNLVFIQFNREADGSLKSLPAKHVDTGMGFERLTSILQ 906
Score = 24.3 bits (51), Expect(2) = 4e-20
Identities = 13/24 (54%), Positives = 14/24 (58%)
Frame = +3
Query: 2 *HPSXWTLHRNSFXQXWQPGCCIF 25
*H S WTL N+F Q W P IF
Sbjct: 687 *HWSLWTLL*NTF*QNW*PQRFIF 758
>BF641925 similar to GP|10177227|db alanyl-tRNA synthetase {Arabidopsis
thaliana}, partial (13%)
Length = 618
Score = 75.9 bits (185), Expect = 2e-14
Identities = 57/183 (31%), Positives = 88/183 (47%), Gaps = 2/183 (1%)
Frame = +3
Query: 175 VANVLGDVFPELKQQEVHIRNVIQEEEESFGRTLVKGIAKFKTAVGHVQGNILSGEATFE 234
V N + ELK++E+ ++ E+ LV I + + G V L+GE F
Sbjct: 102 VKNRAPRILEELKREELRFVQTLERGEKLLEEKLVDAITSAERS-GTVP--CLAGEDVFI 272
Query: 235 LWDTYGFPLDLTQLMAEEKKLKVDVEGFNSAMEAARERSRTAQN--KQAGGAIVMDADAT 292
L+DTYGFP+++T+ +AEE+ + +D+ GF+ ME R +S+ A N K A G+ AD
Sbjct: 273 LYDTYGFPMEITKEVAEERGVSIDMNGFDIEMEKQRRQSQAAHNTVKLAIGSGTNIADN- 449
Query: 293 SALQKRSIVPTDDSFKFASFRDHETVVKAIYTGSEFVDSVNTDGDVGVILESTSFYAEQG 352
VP + + S V + G V V +V ++L T FYAE
Sbjct: 450 --------VPDTEFIGYESLYCKAIVASLVVNGDPXV-QVXEGSNVEILLNXTPFYAESX 602
Query: 353 GQV 355
GQ+
Sbjct: 603 GQI 611
>TC78318 similar to PIR|T45852|T45852 hypothetical protein F3A4.70 -
Arabidopsis thaliana, partial (48%)
Length = 1923
Score = 33.9 bits (76), Expect = 0.10
Identities = 24/56 (42%), Positives = 30/56 (52%)
Frame = +1
Query: 153 AVRYGREVLKAEEGFFNGLVNVVANVLGDVFPELKQQEVHIRNVIQEEEESFGRTL 208
A R+G +V EE F V AN+ DV E +EVH+RN + EES RTL
Sbjct: 589 ASRFGSDVEDLEEDFV-----VKANLCEDVDDE---EEVHVRNGMNFTEESMNRTL 732
>BQ147259 similar to PIR|C96654|C96 hypothetical protein F16P17.10 [imported]
- Arabidopsis thaliana, partial (16%)
Length = 662
Score = 28.9 bits (63), Expect = 3.4
Identities = 28/125 (22%), Positives = 51/125 (40%), Gaps = 8/125 (6%)
Frame = +2
Query: 211 GIAKFKTAVGHVQGNILSGEATFELWDTYGFPLDLTQLMAEEKKLK--------VDVEGF 262
G+ KF AVG+V + E D Y F + L +L+ K ++ V E
Sbjct: 77 GLTKFHNAVGYVAPELAQSFRQSEKCDVYSFGVILLELVTGRKPVESVTAHEVVVLCEYV 256
Query: 263 NSAMEAARERSRTAQNKQAGGAIVMDADATSALQKRSIVPTDDSFKFASFRDHETVVKAI 322
S +E + +N Q ++ + ++ I ++D + S + V+++I
Sbjct: 257 RSLLETGSASNCFDRNLQG----FVENELIQVMKLGLICTSEDPLRRPSMAEIVQVLESI 424
Query: 323 YTGSE 327
GSE
Sbjct: 425 RDGSE 439
>TC90404 similar to GP|21593619|gb|AAM65586.1 receptor protein kinase-like
protein {Arabidopsis thaliana}, partial (52%)
Length = 1252
Score = 28.5 bits (62), Expect = 4.4
Identities = 21/81 (25%), Positives = 40/81 (48%)
Frame = +3
Query: 176 ANVLGDVFPELKQQEVHIRNVIQEEEESFGRTLVKGIAKFKTAVGHVQGNILSGEATFEL 235
AN+L D + E + + ++ + ++S T V+G VGH+ LS + E
Sbjct: 468 ANILLDNYYEAVVGDFGLAKLL-DHQDSHVTTAVRG------TVGHIAPEYLSTGQSSEK 626
Query: 236 WDTYGFPLDLTQLMAEEKKLK 256
D +GF + L +L+ ++ L+
Sbjct: 627 TDVFGFGILLLELITGQRALE 689
>BQ148646 weakly similar to GP|11596186|gb| cystatin-like protein {Citrus x
paradisi}, partial (34%)
Length = 645
Score = 28.1 bits (61), Expect = 5.7
Identities = 12/38 (31%), Positives = 23/38 (59%)
Frame = +2
Query: 16 QXWQPGCCIFGNNDDPTCIEIWNLVLXQFNREADSSLK 53
Q + G GN +DP IEI N + +F++++ +++K
Sbjct: 68 QAIEDGYSPIGNINDPHVIEIANFAVTEFDKKSGATIK 181
>AW329855 similar to PIR|T51383|T51 receptor protein kinase-like protein -
Arabidopsis thaliana, partial (26%)
Length = 577
Score = 28.1 bits (61), Expect = 5.7
Identities = 21/81 (25%), Positives = 40/81 (48%)
Frame = +1
Query: 176 ANVLGDVFPELKQQEVHIRNVIQEEEESFGRTLVKGIAKFKTAVGHVQGNILSGEATFEL 235
AN+L D + E + + ++ + +S T V+G VGH+ LS + E
Sbjct: 181 ANILLDDYCEAVVGDFGLAKLL-DHRDSHVTTAVRG------TVGHIAPEYLSTGQSSEK 339
Query: 236 WDTYGFPLDLTQLMAEEKKLK 256
D +GF + L +L++ ++ L+
Sbjct: 340 TDVFGFGILLLELISGQRALE 402
>TC76782 similar to GP|12539609|gb|AAG59584.1 pathogen-inducible
alpha-dioxygenase {Nicotiana attenuata}, partial (18%)
Length = 390
Score = 28.1 bits (61), Expect = 5.7
Identities = 14/37 (37%), Positives = 19/37 (50%)
Frame = +3
Query: 214 KFKTAVGHVQGNILSGEATFELWDTYGFPLDLTQLMA 250
KFK GH +ILSG + + +G P LT+ A
Sbjct: 30 KFKDTFGHSGNSILSGFVGMKRAENHGVPYSLTEEFA 140
>TC77075 similar to GP|21593294|gb|AAM65243.1 putative carboxyl-terminal
peptidase {Arabidopsis thaliana}, partial (76%)
Length = 2015
Score = 28.1 bits (61), Expect = 5.7
Identities = 20/84 (23%), Positives = 39/84 (45%)
Frame = +1
Query: 126 TLSFAIADGSRPGNEGREYVLRRILRRAVRYGREVLKAEEGFFNGLVNVVANVLGDVFPE 185
T+S +G G ++ +++ L+ R + +K+ +G V+V D PE
Sbjct: 328 TISTTTGNGGNSGGSIQKLEVKKHLKNLNRPPVKSIKSPDGDIIDCVHVSHQPAFD-HPE 504
Query: 186 LKQQEVHIRNVIQEEEESFGRTLV 209
LK ++ +R E ++FG + V
Sbjct: 505 LKDHKIQMRPNFHPERKTFGESKV 576
>TC76781 similar to GP|12539609|gb|AAG59584.1 pathogen-inducible
alpha-dioxygenase {Nicotiana attenuata}, partial (94%)
Length = 2232
Score = 28.1 bits (61), Expect = 5.7
Identities = 14/37 (37%), Positives = 19/37 (50%)
Frame = +2
Query: 214 KFKTAVGHVQGNILSGEATFELWDTYGFPLDLTQLMA 250
KFK GH +ILSG + + +G P LT+ A
Sbjct: 1097 KFKDTFGHSGNSILSGFVGMKRAENHGVPYSLTEEFA 1207
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,135,896
Number of Sequences: 36976
Number of extensions: 104835
Number of successful extensions: 503
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 500
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 501
length of query: 374
length of database: 9,014,727
effective HSP length: 98
effective length of query: 276
effective length of database: 5,391,079
effective search space: 1487937804
effective search space used: 1487937804
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144765.18


