

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144759.6 + phase: 0
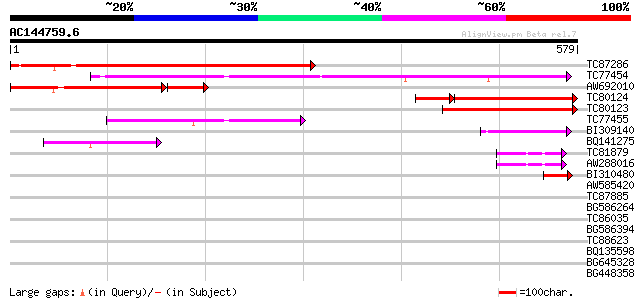
(579 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87286 similar to GP|22335707|dbj|BAC10552. nine-cis-epoxycarot... 490 e-139
TC77454 homologue to GP|22335695|dbj|BAC10549. nine-cis-epoxycar... 335 2e-92
AW692010 similar to GP|22335707|db nine-cis-epoxycarotenoid diox... 263 2e-87
TC80124 homologue to GP|22335707|dbj|BAC10552. nine-cis-epoxycar... 242 3e-78
TC80123 similar to GP|22335707|dbj|BAC10552. nine-cis-epoxycarot... 260 1e-69
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 153 2e-37
BI309140 similar to GP|22335695|db nine-cis-epoxycarotenoid diox... 70 2e-12
BQ141275 similar to GP|22335695|dbj nine-cis-epoxycarotenoid dio... 62 8e-10
TC81879 similar to GP|21902049|dbj|BAC05598. hypothetical protei... 58 9e-09
AW288016 similar to GP|21902049|db hypothetical protein~similar ... 58 9e-09
BI310480 homologue to GP|22335703|dbj nine-cis-epoxycarotenoid d... 42 6e-04
AW585420 weakly similar to GP|21902049|dbj hypothetical protein~... 34 0.14
TC87885 ankyrin-kinase [Medicago truncatula] 33 0.30
BG586264 similar to PIR|T49193|T491 neoxanthin cleavage enzyme n... 31 1.5
TC86035 similar to PIR|E71435|E71435 probable triacylglycerol li... 30 2.6
BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophi... 29 4.4
TC88623 similar to PIR|C84609|C84609 hypothetical protein At2g22... 29 4.4
BQ135598 29 5.7
BG645328 29 5.7
BG448358 28 9.7
>TC87286 similar to GP|22335707|dbj|BAC10552. nine-cis-epoxycarotenoid
dioxygenase4 {Pisum sativum}, partial (50%)
Length = 987
Score = 490 bits (1262), Expect = e-139
Identities = 252/319 (78%), Positives = 271/319 (83%), Gaps = 7/319 (2%)
Frame = +1
Query: 1 MVPKPIIIIACKQPPTTY-IPQFNTASVRTKEKPQTSQPKTLRTT----PTKTLLALLT- 54
MVPKPIII + KQPPTT IP FN +S++TKEKPQTSQ +TL+TT PTKTL T
Sbjct: 46 MVPKPIIITS-KQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTI 222
Query: 55 TTTQKLKNTNKTLQTSSISALILNTLDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELP 114
TTT LK QTSS+ AL+ NT DDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELP
Sbjct: 223 TTTHNLKKP----QTSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELP 390
Query: 115 PTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHAVKITNGKATLCSRYV 174
PTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHA+ I+NGKATLCSRYV
Sbjct: 391 PTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHAITISNGKATLCSRYV 570
Query: 175 QTYKYKIENEAGYQILPNVFSGFNSLIGLAARGSVAVARVLTGQYNPINGIGLINTSLAL 234
QTYKYKIENEAGYQ++PNVFSGFNSLI AARGSV ARV++GQYNP NGIGL NTSLAL
Sbjct: 571 QTYKYKIENEAGYQLIPNVFSGFNSLIASAARGSVTAARVISGQYNPSNGIGLANTSLAL 750
Query: 235 FGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGKLFMNMTAHPKIDSDTGECFAYRYG 294
FGN+LFALGESDLPYEI +TPNGDIQTIGRYDFNGKL M+MTAHPKID DTGE FA+RYG
Sbjct: 751 FGNKLFALGESDLPYEINLTPNGDIQTIGRYDFNGKLSMSMTAHPKIDGDTGETFAFRYG 930
Query: 295 LIRPFLTYFR-FDSNGVKH 312
+ PF F NGVKH
Sbjct: 931 PMPPFFNLFSVLMPNGVKH 987
>TC77454 homologue to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (98%)
Length = 1959
Score = 335 bits (860), Expect = 2e-92
Identities = 196/512 (38%), Positives = 286/512 (55%), Gaps = 21/512 (4%)
Frame = +2
Query: 83 IINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQF 142
I+ F D + P H L+ NFAPV DE PP + IKG LP LNG ++R GPN +F
Sbjct: 176 IVKLFYDSSL-----PHHWLAGNFAPVKDETPPIKDLPIKGHLPDCLNGEFVRVGPNSKF 340
Query: 143 LPRGPYHLFDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNV--FSGFNSL 200
P YH FDGDGM+H ++I +GKAT SR+V+T + K E G + G L
Sbjct: 341 APVAGYHWFDGDGMIHGLRIKDGKATYVSRFVKTSRLKQEEYFGGSKFMKIGDMKGLFGL 520
Query: 201 IGLAARGSVAVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQ 260
+ + + A +V+ Y G G NT+L +L AL E D PY IKV +GD+Q
Sbjct: 521 LMVNMQMLRAKLKVVDVSY----GHGTANTALVYHHQKLLALSEGDKPYAIKVFEDGDLQ 688
Query: 261 TIGRYDFNGKLFMNMTAHPKIDSDTGECFAYRYGLIRPFLTYFRFDSNGVKHNDVPVFSM 320
T+G D++ +L N TAHPK+D TGE F + Y P++TY +G H+ VP+ ++
Sbjct: 689 TLGMLDYDKRLGHNFTAHPKVDPFTGEMFTFGYSHTPPYITYRVISKDGFMHDPVPI-TI 865
Query: 321 KRPSFLHDFAITKKYALFADIQLEINPLDMIFGGSPVRS-DPSKVPRIGILPRYADNESK 379
P +HDFAIT+ Y++F D+ L P +M+ + + S D +K R G+LPRYA +E
Sbjct: 866 SEPIMMHDFAITENYSIFMDLPLYFRPKEMVKNKTLIFSFDSTKKARFGVLPRYAKDEKH 1045
Query: 380 MKWFDVPGLNIMHLINAWDEEDG---KTVTIVAPNILCV-EHMMERLELIHEMIEKVRIN 435
++WF++P I H NAW+EED T + PN+ V + E+L+ + ++R N
Sbjct: 1046IRWFELPNCFIFHNANAWEEEDEIVLITCRLENPNLDMVGGAVKEKLDNFANELYEMRFN 1225
Query: 436 VETGIVTRQPLSARNLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDV-------- 487
++TG +++ LSA +D +N + G+K R++Y + + K +G+VK D+
Sbjct: 1226MKTGEASQKKLSASAVDFPRVNESYTGRKQRYVYGTELDSIAKVTGIVKFDLHAKPDSGK 1405
Query: 488 ----LKGEEVGCRLYGEDCYGGEPFFVAREDG--EEEDDGYLVSYVHDEKKGESRFLVMD 541
+ G VG G +G E +V R G EEDDGYL+ +VHDE G+S V+D
Sbjct: 1406TKLEVGGNVVGHYDLGPGRFGSEAVYVPRVPGTDSEEDDGYLIFFVHDENTGKSFVHVLD 1585
Query: 542 AKSPEFEIVAEVKLPRRVPYGFHGLFVKESDI 573
AK+ + VA V+LP+RVPYGFH FV E +
Sbjct: 1586AKTMSADPVAVVELPQRVPYGFHAFFVTEEQL 1681
>AW692010 similar to GP|22335707|db nine-cis-epoxycarotenoid dioxygenase4
{Pisum sativum}, partial (21%)
Length = 653
Score = 263 bits (672), Expect(2) = 2e-87
Identities = 136/164 (82%), Positives = 142/164 (85%), Gaps = 4/164 (2%)
Frame = +2
Query: 1 MVPKPIIIIACKQPPTTYIPQFNTASVRTKEKPQTSQP-KTLRT---TPTKTLLALLTTT 56
MVPKPIII+ KQPPT IPQFN A VRT+EKPQTSQP +TLRT TKTLL T
Sbjct: 26 MVPKPIIILTTKQPPTIPIPQFNIAFVRTQEKPQTSQPHETLRTGSPAKTKTLL-----T 190
Query: 57 TQKLKNTNKTLQTSSISALILNTLDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPT 116
+QK KNTNK LQTSSISALILNTLDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPT
Sbjct: 191 SQKHKNTNKRLQTSSISALILNTLDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPT 370
Query: 117 QCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHAV 160
QC +IKGTLPPSL G YIRNGPNPQFLPRGPYHLFDGDGMLHA+
Sbjct: 371 QCVIIKGTLPPSLYGAYIRNGPNPQFLPRGPYHLFDGDGMLHAI 502
Score = 78.6 bits (192), Expect(2) = 2e-87
Identities = 35/42 (83%), Positives = 41/42 (97%)
Frame = +3
Query: 162 ITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGL 203
I+NGKATLCSRYV+TYKYKIEN+AG+Q++PNVFSGFNSLI L
Sbjct: 507 ISNGKATLCSRYVKTYKYKIENKAGHQLIPNVFSGFNSLIAL 632
>TC80124 homologue to GP|22335707|dbj|BAC10552. nine-cis-epoxycarotenoid
dioxygenase4 {Pisum sativum}, partial (28%)
Length = 758
Score = 242 bits (618), Expect(2) = 3e-78
Identities = 115/125 (92%), Positives = 122/125 (97%)
Frame = +3
Query: 455 VINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGEDCYGGEPFFVARED 514
VIN +++GKK+RF+YAAIGNPMPK SGVVKIDVLKGEEVGCRLYGE CYGGEPFFVARED
Sbjct: 126 VINGDYMGKKNRFVYAAIGNPMPKISGVVKIDVLKGEEVGCRLYGEGCYGGEPFFVARED 305
Query: 515 GEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFVKESDIT 574
GEEEDDGYLVSYVHDEKKGES+FLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFVKESDIT
Sbjct: 306 GEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFVKESDIT 485
Query: 575 RLSVS 579
+LSVS
Sbjct: 486 KLSVS 500
Score = 68.6 bits (166), Expect(2) = 3e-78
Identities = 31/40 (77%), Positives = 37/40 (92%)
Frame = +2
Query: 415 VEHMMERLELIHEMIEKVRINVETGIVTRQPLSARNLDLA 454
+EH MERLEL+H MIEKV+IN++TGIV+RQPLSARNLD A
Sbjct: 5 IEHTMERLELVHAMIEKVKINIKTGIVSRQPLSARNLDFA 124
>TC80123 similar to GP|22335707|dbj|BAC10552. nine-cis-epoxycarotenoid
dioxygenase4 {Pisum sativum}, partial (23%)
Length = 657
Score = 260 bits (664), Expect = 1e-69
Identities = 126/137 (91%), Positives = 131/137 (94%)
Frame = +3
Query: 443 RQPLSARNLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGEDC 502
R PLSARNLDLAVI N+FLGKK RF+YAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGE C
Sbjct: 3 RHPLSARNLDLAVITNDFLGKKSRFVYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGEGC 182
Query: 503 YGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYG 562
YGGEPFFVA E GEEEDDGYLVSYVHDEKKGES+FLVMDAKSPEFEIVAEVKLPRRVPYG
Sbjct: 183 YGGEPFFVAMEGGEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPRRVPYG 362
Query: 563 FHGLFVKESDITRLSVS 579
FHGLFVK+SDIT+LSVS
Sbjct: 363 FHGLFVKQSDITKLSVS 413
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 153 bits (387), Expect = 2e-37
Identities = 85/206 (41%), Positives = 116/206 (56%), Gaps = 3/206 (1%)
Frame = +3
Query: 100 HVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHA 159
H L+ NFAPV DE PP + +KG LP LNG ++R GPNP+F P YH FDGDGM+H
Sbjct: 1251 HWLAGNFAPVQDETPPIKDLPVKGHLPDCLNGEFVRVGPNPKFSPVAGYHWFDGDGMIHG 1430
Query: 160 VKITNGKATLCSRYVQTYKYKIENEAG---YQILPNVFSGFNSLIGLAARGSVAVARVLT 216
++I +GKAT S +V+T + K E G + + +V G L+ + + ++L
Sbjct: 1431 LRIKDGKATYVSHFVRTSRLKQEEYFGGCKFMKIGDV-KGLLGLLMVTIQTLREKLKILD 1607
Query: 217 GQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGKLFMNMT 276
Y G G NT L +L AL E D PY IKV +GD+ T+G D+ +L T
Sbjct: 1608 VSY----GTGTANTGLVYHNGKLLALSERDKPYAIKVFEDGDLHTLGMLDYEKRLDHYFT 1775
Query: 277 AHPKIDSDTGECFAYRYGLIRPFLTY 302
AHPK+D TGE F + Y P++TY
Sbjct: 1776 AHPKVDPFTGEMFTFGYSQTPPYITY 1853
>BI309140 similar to GP|22335695|db nine-cis-epoxycarotenoid dioxygenase1
{Pisum sativum}, partial (18%)
Length = 589
Score = 70.1 bits (170), Expect = 2e-12
Identities = 38/95 (40%), Positives = 56/95 (58%), Gaps = 2/95 (2%)
Frame = +3
Query: 481 GVVKIDVLKGEEVGCRLYGEDCYGGEPFFVAREDG--EEEDDGYLVSYVHDEKKGESRFL 538
G K+++ G G G+ + +P +V R G +EDDGYL+ +VHDE +S
Sbjct: 12 GKTKLEI-GGNVHGIYDLGQGTFCSDPIYVPRVPGTDSDEDDGYLIFFVHDENTRKSFVH 188
Query: 539 VMDAKSPEFEIVAEVKLPRRVPYGFHGLFVKESDI 573
V+DAK+ + VA V+LP+RVPYGFH F+ E +
Sbjct: 189 VIDAKTMSADPVAVVELPQRVPYGFHSFFMTEDQL 293
>BQ141275 similar to GP|22335695|dbj nine-cis-epoxycarotenoid dioxygenase1
{Pisum sativum}, partial (17%)
Length = 595
Score = 61.6 bits (148), Expect = 8e-10
Identities = 41/132 (31%), Positives = 61/132 (46%), Gaps = 11/132 (8%)
Frame = +2
Query: 35 TSQPKTLRTTPTKTLLALLTTTTQKLKNTNKTLQTSSISALILNTLD-----------DI 83
+S+ +LR+ + + TTT K T +++ SI I+ D+
Sbjct: 200 SSRLNSLRSYRGTQVFNISTTTIHSHKPTCVKIKSCSIGGGIVKVEPKPRKCFTSKAVDL 379
Query: 84 INTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFL 143
+ + S H ++ NFAPV DE PPT+ +KG LP LNG ++R GPNP+
Sbjct: 380 LENLVVKFFYDSSLSHHWIAGNFAPVKDETPPTKDLPVKGHLPDCLNGEFVRVGPNPKXX 559
Query: 144 PRGPYHLFDGDG 155
P Y DG G
Sbjct: 560 PVAGYXWLDGXG 595
>TC81879 similar to GP|21902049|dbj|BAC05598. hypothetical protein~similar
to Arabidopsis thaliana chromosome 4 At4g32810, partial
(15%)
Length = 520
Score = 58.2 bits (139), Expect = 9e-09
Identities = 31/71 (43%), Positives = 40/71 (55%)
Frame = +3
Query: 498 YGEDCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPR 557
Y E EPFFV R +EDDG ++S + EK GE LV+D + FE +A K P
Sbjct: 177 YEEGAVPSEPFFVPRPGATKEDDGVVISII-SEKNGEGYALVLDGST--FEEIARAKFPY 347
Query: 558 RVPYGFHGLFV 568
+PYG HG +V
Sbjct: 348 GLPYGLHGCWV 380
>AW288016 similar to GP|21902049|db hypothetical protein~similar to
Arabidopsis thaliana chromosome 4 At4g32810, partial
(17%)
Length = 542
Score = 58.2 bits (139), Expect = 9e-09
Identities = 31/71 (43%), Positives = 40/71 (55%)
Frame = +3
Query: 498 YGEDCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPR 557
Y E EPFFV R +EDDG ++S + EK GE LV+D + FE +A K P
Sbjct: 99 YEEGAVPSEPFFVPRPGATKEDDGVVISII-SEKNGEGYALVLDGST--FEEIARAKFPY 269
Query: 558 RVPYGFHGLFV 568
+PYG HG +V
Sbjct: 270 GLPYGLHGCWV 302
>BI310480 homologue to GP|22335703|dbj nine-cis-epoxycarotenoid dioxygenase3
{Pisum sativum}, partial (5%)
Length = 415
Score = 42.0 bits (97), Expect = 6e-04
Identities = 17/29 (58%), Positives = 22/29 (75%)
Frame = +3
Query: 546 EFEIVAEVKLPRRVPYGFHGLFVKESDIT 574
+ ++ A VKLP RVPYGFHG FV+ D+T
Sbjct: 12 DLKVEATVKLPSRVPYGFHGTFVEAKDLT 98
>AW585420 weakly similar to GP|21902049|dbj hypothetical protein~similar to
Arabidopsis thaliana chromosome 4 At4g32810, partial
(13%)
Length = 551
Score = 34.3 bits (77), Expect = 0.14
Identities = 20/64 (31%), Positives = 33/64 (51%), Gaps = 3/64 (4%)
Frame = +3
Query: 120 VIKGTLPPSLNGVYIRNGPNPQFLPRGPY---HLFDGDGMLHAVKITNGKATLCSRYVQT 176
+++G +P L G Y++NGP + G Y HLFDG L + +G+ R V++
Sbjct: 357 LVQGHIPLWLKGTYLKNGPGMWNI--GDYNFRHLFDGYATLVGLHFEDGRLIAGHRQVES 530
Query: 177 YKYK 180
Y+
Sbjct: 531 QAYQ 542
>TC87885 ankyrin-kinase [Medicago truncatula]
Length = 1605
Score = 33.1 bits (74), Expect = 0.30
Identities = 20/56 (35%), Positives = 33/56 (58%), Gaps = 3/56 (5%)
Frame = -3
Query: 42 RTTPTKTLLALLTTTTQKLKNTNKTLQTS---SISALILNTLDDIINTFIDPPIKP 94
RTTPT T + + T QKL+N N + +S S +A+ +N ++ IN+F+ + P
Sbjct: 601 RTTPTITSIKIGLTAEQKLRNINMSFTSSDMKSSTAIKINAIN--INSFVKQILNP 440
>BG586264 similar to PIR|T49193|T491 neoxanthin cleavage enzyme nc1 -
Arabidopsis thaliana, partial (17%)
Length = 660
Score = 30.8 bits (68), Expect = 1.5
Identities = 12/21 (57%), Positives = 15/21 (71%)
Frame = +2
Query: 129 LNGVYIRNGPNPQFLPRGPYH 149
LNG++ R GPNP+F P YH
Sbjct: 575 LNGIFRRVGPNPKFDPLAGYH 637
Score = 30.4 bits (67), Expect = 2.0
Identities = 14/29 (48%), Positives = 17/29 (58%)
Frame = +2
Query: 98 PRHVLSQNFAPVLDELPPTQCKVIKGTLP 126
P LS NFAP+ DE PP + + G LP
Sbjct: 158 PLRYLSGNFAPLRDETPPVKDLPVHGFLP 244
>TC86035 similar to PIR|E71435|E71435 probable triacylglycerol lipase -
Arabidopsis thaliana, partial (57%)
Length = 1783
Score = 30.0 bits (66), Expect = 2.6
Identities = 20/54 (37%), Positives = 27/54 (49%)
Frame = -1
Query: 26 SVRTKEKPQTSQPKTLRTTPTKTLLALLTTTTQKLKNTNKTLQTSSISALILNT 79
SV T E P+ + TTP TL L+ +++ LKN TL +S IL T
Sbjct: 1453 SVPTCEYPKGVFSSNISTTPLTTLEFLIFSSSSSLKNIPGTLVITSCVFTILRT 1292
>BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophila
melanogaster}, partial (2%)
Length = 734
Score = 29.3 bits (64), Expect = 4.4
Identities = 19/61 (31%), Positives = 29/61 (47%)
Frame = +1
Query: 4 KPIIIIACKQPPTTYIPQFNTASVRTKEKPQTSQPKTLRTTPTKTLLALLTTTTQKLKNT 63
+PI+ ++ PT + T E T++P+T TPTK+ + TTT KN
Sbjct: 205 EPIVQTDSEETPTEKPEKETTTKTTKPEAETTTEPET--NTPTKSETDVPETTTTLEKNQ 378
Query: 64 N 64
N
Sbjct: 379 N 381
>TC88623 similar to PIR|C84609|C84609 hypothetical protein At2g22130
[imported] - Arabidopsis thaliana, partial (24%)
Length = 1960
Score = 29.3 bits (64), Expect = 4.4
Identities = 12/30 (40%), Positives = 17/30 (56%)
Frame = +1
Query: 113 LPPTQCKVIKGTLPPSLNGVYIRNGPNPQF 142
+P CK+ G PP L V + GPNP++
Sbjct: 1459 IPSVYCKITLGNSPPKLTKV-VSTGPNPEW 1545
>BQ135598
Length = 843
Score = 28.9 bits (63), Expect = 5.7
Identities = 15/52 (28%), Positives = 26/52 (49%), Gaps = 6/52 (11%)
Frame = +2
Query: 143 LPRGPYHLFDGDGMLHAVKITN------GKATLCSRYVQTYKYKIENEAGYQ 188
+P+ +H + D ++H ++N ATLC ++QT KY GY+
Sbjct: 389 VPKKRHHA*ESDQLIHL*NLSNIYTCLFYYATLCPAFIQTLKY*YLTHLGYK 544
>BG645328
Length = 770
Score = 28.9 bits (63), Expect = 5.7
Identities = 22/71 (30%), Positives = 30/71 (41%), Gaps = 8/71 (11%)
Frame = -2
Query: 494 GCRLYGEDCYGGEPFFVAR------EDGEEEDDGY--LVSYVHDEKKGESRFLVMDAKSP 545
GC +GE C+G F + + GEE + Y L +E G RF AK
Sbjct: 400 GC*CFGEACFGDYAFALCKSYRWYSSSGEESAETY*CLARI**EEDFGNGRFFCYAAKIA 221
Query: 546 EFEIVAEVKLP 556
E+V+ LP
Sbjct: 220 SVELVS*NLLP 188
>BG448358
Length = 400
Score = 28.1 bits (61), Expect = 9.7
Identities = 11/23 (47%), Positives = 17/23 (73%)
Frame = +3
Query: 519 DDGYLVSYVHDEKKGESRFLVMD 541
D+GYL S + D+K GE+ F+ +D
Sbjct: 267 DEGYLQSMLFDDKMGENVFICID 335
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.140 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,887,138
Number of Sequences: 36976
Number of extensions: 248593
Number of successful extensions: 1163
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1140
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1152
length of query: 579
length of database: 9,014,727
effective HSP length: 101
effective length of query: 478
effective length of database: 5,280,151
effective search space: 2523912178
effective search space used: 2523912178
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144759.6


