

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144731.8 - phase: 0
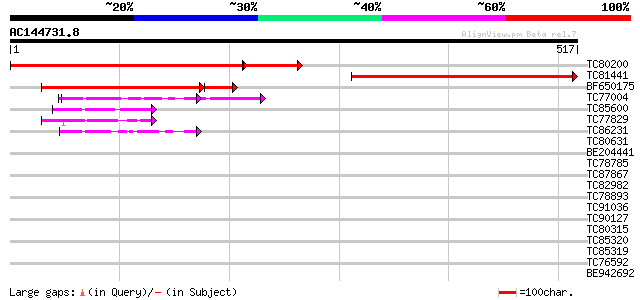
(517 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80200 similar to GP|21928041|gb|AAM78049.1 At2g01270/F10A8.15 ... 444 e-151
TC81441 similar to PIR|F84422|F84422 hypothetical protein At2g01... 436 e-123
BF650175 similar to GP|21928041|gb At2g01270/F10A8.15 {Arabidops... 318 9e-99
TC77004 homologue to SP|P38661|PDA6_MEDSA Probable protein disul... 60 3e-09
TC85600 homologue to SP|P29828|PDI_MEDSA Protein disulfide isome... 51 1e-06
TC77829 similar to GP|7211992|gb|AAF40463.1| Strong simialrity t... 44 2e-04
TC86231 similar to GP|21593313|gb|AAM65262.1 protein disulfide i... 42 7e-04
TC80631 similar to GP|21555659|gb|AAM63908.1 unknown {Arabidopsi... 41 0.001
BE204441 SP|P38661|PDA6 Probable protein disulfide isomerase A6 ... 41 0.001
TC78785 similar to GP|15450830|gb|AAK96686.1 Unknown protein {Ar... 35 0.091
TC87867 similar to GP|15594012|emb|CAC69854. putative thioredoxi... 33 0.20
TC82982 weakly similar to GP|1388082|gb|AAC49355.1| thioredoxin ... 33 0.20
TC78893 similar to GP|16649013|gb|AAL24358.1 Unknown protein {Ar... 33 0.20
TC91036 weakly similar to GP|14423498|gb|AAK62431.1 Unknown prot... 33 0.27
TC90127 similar to PIR|E85439|E85439 thiol-disulfide interchange... 32 0.77
TC80315 similar to PIR|B96796|B96796 thioredoxin-like protein 4... 31 1.0
TC85320 similar to GP|1532175|gb|AAB07885.1| similar to protein ... 31 1.0
TC85319 similar to GP|8778824|gb|AAF79823.1| T6D22.5 {Arabidopsi... 31 1.0
TC76592 similar to GP|1388088|gb|AAC49358.1| thioredoxin m {Pisu... 31 1.0
BE942692 similar to GP|8778824|gb| T6D22.5 {Arabidopsis thaliana... 31 1.0
>TC80200 similar to GP|21928041|gb|AAM78049.1 At2g01270/F10A8.15
{Arabidopsis thaliana}, partial (43%)
Length = 1069
Score = 444 bits (1143), Expect(2) = e-151
Identities = 214/217 (98%), Positives = 215/217 (98%)
Frame = +1
Query: 1 MNLLYFALFLCTISVSSSATSSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDA 60
MNLLYFALFLCTISVSSSATSSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDA
Sbjct: 268 MNLLYFALFLCTISVSSSATSSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDA 447
Query: 61 VLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINI 120
VLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINI
Sbjct: 448 VLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINI 627
Query: 121 KLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADRLLNWINKQMSSSF 180
KLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADRLLNWINKQMSSSF
Sbjct: 628 KLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADRLLNWINKQMSSSF 807
Query: 181 GLDDQKFQNEHLSSNVSDPEQIARAIYDVEEATSLAF 217
GLDDQKFQNEHLSSNVSDPEQIARAIYDVEEAT +F
Sbjct: 808 GLDDQKFQNEHLSSNVSDPEQIARAIYDVEEATFFSF 918
Score = 110 bits (274), Expect(2) = e-151
Identities = 54/54 (100%), Positives = 54/54 (100%)
Frame = +2
Query: 214 SLAFDIILENKMIKPETRASLVKFLQVLTAHHPSRRCRKGAGDLLVSFADLYPT 267
SLAFDIILENKMIKPETRASLVKFLQVLTAHHPSRRCRKGAGDLLVSFADLYPT
Sbjct: 908 SLAFDIILENKMIKPETRASLVKFLQVLTAHHPSRRCRKGAGDLLVSFADLYPT 1069
>TC81441 similar to PIR|F84422|F84422 hypothetical protein At2g01270
[imported] - Arabidopsis thaliana, partial (46%)
Length = 961
Score = 436 bits (1122), Expect = e-123
Identities = 206/206 (100%), Positives = 206/206 (100%)
Frame = +3
Query: 312 CGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVCEECRQHFYDMCSSVSTPFNKARDF 371
CGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVCEECRQHFYDMCSSVSTPFNKARDF
Sbjct: 3 CGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVCEECRQHFYDMCSSVSTPFNKARDF 182
Query: 372 VLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHDQKSNKIEWNQDE 431
VLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHDQKSNKIEWNQDE
Sbjct: 183 VLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHDQKSNKIEWNQDE 362
Query: 432 VYKALKNYYEKTLVSLYKEKDIAGNDGTKGAALEDLIVGTNAVVVPVGAALAIAVASCAF 491
VYKALKNYYEKTLVSLYKEKDIAGNDGTKGAALEDLIVGTNAVVVPVGAALAIAVASCAF
Sbjct: 363 VYKALKNYYEKTLVSLYKEKDIAGNDGTKGAALEDLIVGTNAVVVPVGAALAIAVASCAF 542
Query: 492 GALACYWRSQQKSRKYFHHLHSLKNI 517
GALACYWRSQQKSRKYFHHLHSLKNI
Sbjct: 543 GALACYWRSQQKSRKYFHHLHSLKNI 620
>BF650175 similar to GP|21928041|gb At2g01270/F10A8.15 {Arabidopsis
thaliana}, partial (31%)
Length = 544
Score = 318 bits (816), Expect(2) = 9e-99
Identities = 148/148 (100%), Positives = 148/148 (100%)
Frame = +3
Query: 30 RSILREVHDNDQTGTTDHDYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHY 89
RSILREVHDNDQTGTTDHDYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHY
Sbjct: 3 RSILREVHDNDQTGTTDHDYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHY 182
Query: 90 EKVAKLFNGPDAVHPGIILLTRVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEP 149
EKVAKLFNGPDAVHPGIILLTRVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEP
Sbjct: 183 EKVAKLFNGPDAVHPGIILLTRVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEP 362
Query: 150 KQEKSDIHVIDDARTADRLLNWINKQMS 177
KQEKSDIHVIDDARTADRLLNWINKQMS
Sbjct: 363 KQEKSDIHVIDDARTADRLLNWINKQMS 446
Score = 60.5 bits (145), Expect(2) = 9e-99
Identities = 28/30 (93%), Positives = 28/30 (93%)
Frame = +1
Query: 178 SSFGLDDQKFQNEHLSSNVSDPEQIARAIY 207
SSFGLDDQKFQNEHL SNV DPEQIARAIY
Sbjct: 454 SSFGLDDQKFQNEHLXSNVXDPEQIARAIY 543
>TC77004 homologue to SP|P38661|PDA6_MEDSA Probable protein disulfide
isomerase A6 precursor (EC 5.3.4.1) (P5). [Alfalfa]
{Medicago sativa}, complete
Length = 1575
Score = 59.7 bits (143), Expect = 3e-09
Identities = 40/128 (31%), Positives = 58/128 (45%)
Frame = +2
Query: 48 DYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGII 107
D V L NF+ + A+VEF+A WC C+ P YEK+ F +V
Sbjct: 176 DDVVVLTEENFEKEVGQDKG--ALVEFYAPWCGHCKKLAPEYEKLGNSFKKAKSV----- 334
Query: 108 LLTRVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADR 167
L+ +VDC + C K+ V YP + W P GS EPK+ + RTA+
Sbjct: 335 LIAKVDCDEHKGV--CSKYGVSGYPTIQWFPK-----GSLEPKK-------FEGPRTAES 472
Query: 168 LLNWINKQ 175
L ++N +
Sbjct: 473 LAEFVNTE 496
Score = 45.4 bits (106), Expect = 5e-05
Identities = 47/189 (24%), Positives = 78/189 (40%)
Frame = +2
Query: 45 TDHDYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHP 104
T + V L F+ V+ D +VEF+A WC C++ P YEKVA +F D V
Sbjct: 521 TAPSHVVVLTPETFNEVVLDETKD-VLVEFYAPWCGHCKSLAPIYEKVAAVFKSEDDV-- 691
Query: 105 GIILLTRVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDART 164
++ +D A K L +K+ V +P L + P G + R
Sbjct: 692 ---VIANLD-ADKYR-DLAEKYDVSGFPTLKFFPKGNKAGEDY------------GGGRD 820
Query: 165 ADRLLNWINKQMSSSFGLDDQKFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILENK 224
D + +IN++ +S D K Q + V D +++ + + A +E +
Sbjct: 821 LDDFVAFINEKSGTS---RDAKGQLTSEAGIVEDLDELVKEFVAANDEEKKAVFARIEEE 991
Query: 225 MIKPETRAS 233
+ K + AS
Sbjct: 992 VEKLKGSAS 1018
>TC85600 homologue to SP|P29828|PDI_MEDSA Protein disulfide isomerase
precursor (PDI) (EC 5.3.4.1). [Alfalfa] {Medicago
sativa}, complete
Length = 1915
Score = 50.8 bits (120), Expect = 1e-06
Identities = 28/95 (29%), Positives = 47/95 (49%)
Frame = +2
Query: 40 DQTGTTDHDYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGP 99
+++ T ++ + L+ TNF +K F VVEF+A WC C+ P YEK A +
Sbjct: 122 EESSTDAKEFVLTLDNTNFHDTVKKHD--FIVVEFYAPWCGHCKKLAPEYEKAASIL--- 286
Query: 100 DAVHPGIILLTRVDCASKINIKLCDKFSVGHYPML 134
+ H ++L +VD + N L + V +P +
Sbjct: 287 -STHEPPVVLAKVDANEEHNKDLASENDVKGFPTI 388
Score = 34.3 bits (77), Expect = 0.12
Identities = 33/158 (20%), Positives = 61/158 (37%)
Frame = +2
Query: 44 TTDHDYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVH 103
T + V + T D V K ++EF+A WC C+ P ++VA F
Sbjct: 1169 TNNEPVKVVVGQTLEDIVFKS--GKNVLIEFYAPWCGHCKQLAPILDEVAVSFQS----- 1327
Query: 104 PGIILLTRVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDAR 163
+++ ++D + N D F V YP L++ + + D R
Sbjct: 1328 DADVVIAKLDATA--NDIPTDTFEVQGYPTLYF-------------RSASGKLSQYDGGR 1462
Query: 164 TADRLLNWINKQMSSSFGLDDQKFQNEHLSSNVSDPEQ 201
T + ++ +I K + G Q+ + ++ + EQ
Sbjct: 1463 TKEDIIEFIEKNKDKT-GAAHQEVEQPKAAAAQPEAEQ 1573
>TC77829 similar to GP|7211992|gb|AAF40463.1| Strong simialrity to the
disulfide isomerase precursor homolog T21L14.14
GP|2702281 from A., partial (89%)
Length = 1646
Score = 43.9 bits (102), Expect = 2e-04
Identities = 29/111 (26%), Positives = 56/111 (50%), Gaps = 6/111 (5%)
Frame = +2
Query: 30 RSILREVHDNDQTGTTDH------DYAVELNATNFDAVLKDTPATFAVVEFFANWCPACR 83
+++L+E + TG ++ +VELN++NFD ++ + + +VEFFA WC C+
Sbjct: 428 KALLKERLNGKATGGSNEKKESTASSSVELNSSNFDELVIKSKELW-IVEFFAPWCGHCK 604
Query: 84 NYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINIKLCDKFSVGHYPML 134
P +++ + G + L VDC + + L +F+V +P +
Sbjct: 605 KLAPEWKRASNNLKGK-------VKLGHVDCDA--DKSLMSRFNVQGFPTI 730
Score = 33.9 bits (76), Expect = 0.16
Identities = 16/48 (33%), Positives = 26/48 (53%)
Frame = +2
Query: 51 VELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNG 98
++L NF + + ++ +VEFFA WC C+ P +EK A + G
Sbjct: 119 LQLTPNNFKSKVLNSNGV-VLVEFFAPWCGHCKALTPIWEKAATVLKG 259
>TC86231 similar to GP|21593313|gb|AAM65262.1 protein disulfide isomerase
precursor-like {Arabidopsis thaliana}, partial (79%)
Length = 2185
Score = 41.6 bits (96), Expect = 7e-04
Identities = 33/130 (25%), Positives = 59/130 (45%)
Frame = +1
Query: 46 DHDYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPG 105
D V L NF V+++ F +VEF+A WC C+ P Y A + G
Sbjct: 352 DEKDVVVLIERNFTTVIENNQ--FVMVEFYAPWCGHCQALAPEYAAAA-----TELKKDG 510
Query: 106 IILLTRVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTA 165
++ L +VD + + +L +++V +P +++ F+ G +P + RT
Sbjct: 511 VV-LAKVD--ASVENELAYEYNVQGFPTVYF-----FIDGVHKP---------YNGQRTK 639
Query: 166 DRLLNWINKQ 175
D ++ WI K+
Sbjct: 640 DAIVTWIKKK 669
>TC80631 similar to GP|21555659|gb|AAM63908.1 unknown {Arabidopsis
thaliana}, partial (60%)
Length = 1296
Score = 41.2 bits (95), Expect = 0.001
Identities = 21/77 (27%), Positives = 34/77 (43%), Gaps = 3/77 (3%)
Frame = +2
Query: 315 WVLLHSLSVRIEDGESQFTFNAICDFVH---NFFVCEECRQHFYDMCSSVSTPFNKARDF 371
W LH+L+ + D ++ + + V + C+EC HF ++ S +F
Sbjct: 749 WTFLHTLAAQYPDNPTRQQKKDVKELVQILSRMYPCKECADHFKEVLRSNPVQAGSHAEF 928
Query: 372 VLWLWSSHNKVNERLSK 388
WL HN VN +SK
Sbjct: 929 SQWLCHVHNVVNRSISK 979
>BE204441 SP|P38661|PDA6 Probable protein disulfide isomerase A6 precursor
(EC 5.3.4.1) (P5). [Alfalfa] {Medicago sativa}, partial
(38%)
Length = 416
Score = 41.2 bits (95), Expect = 0.001
Identities = 22/58 (37%), Positives = 30/58 (50%)
Frame = +3
Query: 45 TDHDYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAV 102
T + V L F+ V+ D +VEF+A WC C++ P YEKVA +F D V
Sbjct: 183 TAPSHVVVLTPETFNEVVLDETKD-VLVEFYAPWCGHCKSLAPIYEKVAAVFKSEDDV 353
>TC78785 similar to GP|15450830|gb|AAK96686.1 Unknown protein {Arabidopsis
thaliana}, partial (57%)
Length = 1477
Score = 34.7 bits (78), Expect = 0.091
Identities = 20/62 (32%), Positives = 31/62 (49%), Gaps = 2/62 (3%)
Frame = -2
Query: 357 MCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTK--QLCPSC 414
+CS T ++ F+LW+W +V R+S+ + S G+ K+IWPT CP
Sbjct: 1386 ICSIRITAYDVLNIFILWIW---KEVTIRISRLKFS*GS*SQDTTKSIWPTSAFDTCPEA 1216
Query: 415 YL 416
L
Sbjct: 1215 KL 1210
>TC87867 similar to GP|15594012|emb|CAC69854. putative thioredoxin m2 {Pisum
sativum}, partial (79%)
Length = 816
Score = 33.5 bits (75), Expect = 0.20
Identities = 27/95 (28%), Positives = 48/95 (50%), Gaps = 1/95 (1%)
Frame = +3
Query: 5 YFALFLCTISVSSSATSSSSSSFP-GRSILREVHDNDQTGTTDHDYAVELNATNFDAVLK 63
Y L L +S + + S+ FP G +++ E D TT + A + N+ +++
Sbjct: 141 YTGLKLRPVSATRLRSQSTGRVFPRGGTVVCEARD-----TTAVEVA-SITDGNWQSLVI 302
Query: 64 DTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNG 98
++ T +VEF+A WC CR P +++AK + G
Sbjct: 303 ESE-TPVLVEFWAPWCGPCRMIHPIIDELAKEYAG 404
>TC82982 weakly similar to GP|1388082|gb|AAC49355.1| thioredoxin h
{Arabidopsis thaliana}, partial (38%)
Length = 481
Score = 33.5 bits (75), Expect = 0.20
Identities = 20/89 (22%), Positives = 39/89 (43%), Gaps = 9/89 (10%)
Frame = +1
Query: 57 NFDAVLKDTPATF---------AVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGII 107
NF + KD F +V+F A+WCP C+ P ++ +A+ + +
Sbjct: 28 NFIVIQKDNKDDFNKELNKGKLVIVDFTASWCPPCKMIAPIFKNLAEEYK-------KVA 186
Query: 108 LLTRVDCASKINIKLCDKFSVGHYPMLFW 136
+ +VD N ++ ++ + YP F+
Sbjct: 187 IFLKVD--GDENPEITKEYGISAYPTFFF 267
>TC78893 similar to GP|16649013|gb|AAL24358.1 Unknown protein {Arabidopsis
thaliana}, partial (32%)
Length = 805
Score = 33.5 bits (75), Expect = 0.20
Identities = 19/67 (28%), Positives = 32/67 (47%)
Frame = +3
Query: 69 FAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINIKLCDKFSV 128
+ V F+A+WCP R ++P +F+G ++HP I + S + K+ V
Sbjct: 576 YVAVLFYASWCPFSRIFRP-------VFSGISSLHPTIPHFAIEE--SSVRPSTLSKYGV 728
Query: 129 GHYPMLF 135
+P LF
Sbjct: 729 HGFPTLF 749
>TC91036 weakly similar to GP|14423498|gb|AAK62431.1 Unknown protein
{Arabidopsis thaliana}, partial (42%)
Length = 744
Score = 33.1 bits (74), Expect = 0.27
Identities = 32/135 (23%), Positives = 53/135 (38%), Gaps = 3/135 (2%)
Frame = +2
Query: 53 LNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRV 112
L+ +NFD+ + +V+F+A WC C+ P + A + A IL+ +V
Sbjct: 140 LDESNFDSAISSFD--HILVDFYAPWCGHCKRLSPELDAAAPVL----AALKEPILIAKV 301
Query: 113 DCASKINIKLCDKFSVGHYPMLF---WGPPPKFVGGSWEPKQEKSDIHVIDDARTADRLL 169
D + L K V YP + G P ++ G R AD L+
Sbjct: 302 DADK--HTSLARKHDVDAYPTILLFNHGVPTEYRG-----------------PRKADLLV 424
Query: 170 NWINKQMSSSFGLDD 184
++ K +S + D
Sbjct: 425 RYLKKFAASDVSILD 469
>TC90127 similar to PIR|E85439|E85439 thiol-disulfide interchange like
protein [imported] - Arabidopsis thaliana, partial (53%)
Length = 754
Score = 31.6 bits (70), Expect = 0.77
Identities = 11/26 (42%), Positives = 16/26 (61%)
Frame = +1
Query: 71 VVEFFANWCPACRNYKPHYEKVAKLF 96
VVEF+A+WC CR P K+ + +
Sbjct: 490 VVEFYADWCEVCRELAPDVYKIEQQY 567
>TC80315 similar to PIR|B96796|B96796 thioredoxin-like protein 49720-48645
[imported] - Arabidopsis thaliana, partial (70%)
Length = 802
Score = 31.2 bits (69), Expect = 1.0
Identities = 22/82 (26%), Positives = 40/82 (47%), Gaps = 3/82 (3%)
Frame = +2
Query: 15 VSSSATSSSSSSFPGRSI---LREVHDNDQTGTTDHDYAVELNATNFDAVLKDTPATFAV 71
V S++SSS FP R + R + + ++ A + ++FD +L ++
Sbjct: 50 VGPSSSSSSQLQFPVRLLPIGTRRLSSSSRSRFLPLVQAKKQTFSSFDDLLANSDKP-VF 226
Query: 72 VEFFANWCPACRNYKPHYEKVA 93
V+F+A WC C+ P E+V+
Sbjct: 227 VDFYATWCGPCQFMVPVLEEVS 292
>TC85320 similar to GP|1532175|gb|AAB07885.1| similar to protein disulfide
isomerase {Arabidopsis thaliana}, partial (78%)
Length = 746
Score = 31.2 bits (69), Expect = 1.0
Identities = 19/82 (23%), Positives = 34/82 (41%)
Frame = +1
Query: 51 VELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLT 110
+ L + F +K+ + V+F WC C+N ++ V K + + G
Sbjct: 151 ITLTSDTFSDKIKEKDTAW-FVKFCVPWCKYCKNLGSLWDDVGKAMENENEIEIG----- 312
Query: 111 RVDCASKINIKLCDKFSVGHYP 132
VDC + + +C K + YP
Sbjct: 313 EVDCGT--DKAVCSKVDIHSYP 372
>TC85319 similar to GP|8778824|gb|AAF79823.1| T6D22.5 {Arabidopsis
thaliana}, partial (62%)
Length = 881
Score = 31.2 bits (69), Expect = 1.0
Identities = 19/82 (23%), Positives = 34/82 (41%)
Frame = +1
Query: 51 VELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLT 110
+ L + F +K+ + V+F WC C+N ++ V K + + G
Sbjct: 202 ITLTSDTFSDKIKEKDTAW-FVKFCVPWCKYCKNLGSLWDDVGKAMENENEIEIG----- 363
Query: 111 RVDCASKINIKLCDKFSVGHYP 132
VDC + + +C K + YP
Sbjct: 364 EVDCGT--DKAVCSKVDIHSYP 423
>TC76592 similar to GP|1388088|gb|AAC49358.1| thioredoxin m {Pisum sativum},
partial (87%)
Length = 912
Score = 31.2 bits (69), Expect = 1.0
Identities = 17/49 (34%), Positives = 27/49 (54%)
Frame = +1
Query: 50 AVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNG 98
AV ++ N + DTP +V+F+A WC CR P +++AK + G
Sbjct: 325 AVTDSSWNELVLASDTPV---LVDFWAPWCGPCRMIAPIIDELAKEYAG 462
>BE942692 similar to GP|8778824|gb| T6D22.5 {Arabidopsis thaliana}, partial
(62%)
Length = 588
Score = 31.2 bits (69), Expect = 1.0
Identities = 19/82 (23%), Positives = 34/82 (41%)
Frame = +1
Query: 51 VELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLT 110
+ L + F +K+ + V+F WC C+N ++ V K + + G
Sbjct: 112 ITLTSDTFSDKIKEKDTAW-FVKFCVPWCKYCKNLGSLWDDVGKAMENENEIEIG----- 273
Query: 111 RVDCASKINIKLCDKFSVGHYP 132
VDC + + +C K + YP
Sbjct: 274 EVDCGT--DKAVCSKVDIHSYP 333
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.135 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,140,307
Number of Sequences: 36976
Number of extensions: 334465
Number of successful extensions: 2257
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 2050
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2205
length of query: 517
length of database: 9,014,727
effective HSP length: 100
effective length of query: 417
effective length of database: 5,317,127
effective search space: 2217241959
effective search space used: 2217241959
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144731.8


