

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144731.14 + phase: 0 /pseudo
(1343 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
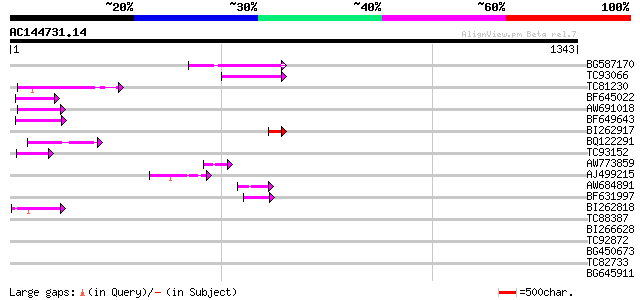
Score E
Sequences producing significant alignments: (bits) Value
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 174 3e-43
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 121 2e-27
TC81230 100 5e-21
BF645022 weakly similar to GP|8777581|dbj retroelement pol polyp... 62 1e-09
AW691018 weakly similar to GP|2462935|em open reading frame 1 {B... 57 5e-08
BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element pol... 54 5e-07
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 53 9e-07
BQ122291 52 1e-06
TC93152 50 6e-06
AW773859 49 1e-05
AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 47 6e-05
AW684891 47 6e-05
BF631997 weakly similar to GP|18542925|gb Putative pol polyprote... 44 3e-04
BI262818 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 44 4e-04
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 42 0.001
BI266628 40 0.006
TC92872 38 0.023
BG450673 homologue to GP|12055499|emb NADH dehydrogenase subunit... 35 0.080
TC82733 similar to GP|10177404|dbj|BAB10535. gene_id:K24M7.12~pi... 36 0.11
BG645911 35 0.15
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 174 bits (440), Expect = 3e-43
Identities = 93/238 (39%), Positives = 137/238 (57%), Gaps = 4/238 (1%)
Frame = -3
Query: 423 SGKVIAKGPKVGRLFPLQFISSHLSLACN----NVLNSYEDWHRKLGHPNSTVLSHLFKT 478
S ++I +G G L+ L+ + + C+ + LN WH +LGHP+ L+ +
Sbjct: 704 SSQLIGEGVTKGDLYMLEKLDPVSNYKCSFTSSSSLNKDALWHARLGHPHGRALNLMLPG 525
Query: 479 GLLGNKQVVCTASISCPVCKLAKSKTLPFPSGAHRASNCFEMIHSDVWGMSPIASHAHYK 538
+ NK +C C L K FP + NCF++I++D+W +P S ++K
Sbjct: 524 VVFENK--------NCEACILGKHCKNVFPRTSTVYENCFDLIYTDLW-TAPSLSRDNHK 372
Query: 539 YFVTFIDDYSRFTWIYFLRSKSEVFSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQ 598
YFVTFID+ S++TW+ + SK V FK F YV + A +KI RS++GGEY S+ F+
Sbjct: 371 YFVTFIDEKSKYTWLTLIPSKDRVIDAFKNFQAYVTNHYHAKIKILRSDNGGEYTSYAFK 192
Query: 599 EYLQHKGILSQRSCPNTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFWVEALSTVCF 656
+L H GIL Q SCP TPQQNG+A+RKN+HL++V RSL+ QA+V ST C+
Sbjct: 191 SHLDHHGILHQTSCPYTPQQNGVAKRKNKHLMEVARSLMFQANV---------STACY 45
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 121 bits (303), Expect = 2e-27
Identities = 63/155 (40%), Positives = 89/155 (56%), Gaps = 2/155 (1%)
Frame = +1
Query: 503 KTLPFPSGAHRASNCFEMIHSDVWGMSPIASHAHYKYFVTFIDDYSRFTWIYFLRSKSEV 562
K + F + HR + IHSD+WG S + S+ +Y +T IDD+ R W+YFLR K+E
Sbjct: 52 KKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNET 231
Query: 563 FSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQEYLQHKGILSQRSCPNTPQQNGLA 622
F FKK+ VETQ +VK +++ E+ S +F E+ + GI ++ P PQQNG+A
Sbjct: 232 FPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVA 411
Query: 623 ERKNRHLLDVTRSLLLQASV--PPRFWVEALSTVC 655
ER R LL+ R +L A + WVEA ST C
Sbjct: 412 ERMIRTLLERARCMLSNAGL*N*RDLWVEAASTAC 516
>TC81230
Length = 958
Score = 100 bits (248), Expect = 5e-21
Identities = 68/263 (25%), Positives = 117/263 (43%), Gaps = 10/263 (3%)
Frame = +1
Query: 18 GKNYPAWEFQFRMYVKGNKLWSHLDDVSKAPTEKA---------ALEEWEYKDAQIISWI 68
G NY W ++KG +LW ++ K PT+ LEEW+ K+ QII+W
Sbjct: 244 GSNYNHWAESMCGFLKGRRLWRYVTGDKKCPTKGKDDTADAFADKLEEWDSKNHQIITWF 423
Query: 69 LSSIDPQMINNLRSFSTAQEMWNYLKRIYNQDNAAKRFQLELEIANYK-QGNLYVQEFYS 127
++ P + F A+E+W++LK+ Y + + ++QL +++N K Q V EF +
Sbjct: 424 RNTSIPSIHMQFGRFENAKEVWDHLKQRYTISDLSHQYQLLKDLSNLKQQSGQPVYEFLA 603
Query: 128 GFLNLWTEHSAIIHADVPKASLAAVQEVYNTSRRDQFLMKLRPEFEVVRGALLNRNPVPS 187
+W + ++ + + + N R QFLM L E+E VR + L++NP+P+
Sbjct: 604 QMEVIWNQLTSCEPSLKDATDMKTYETHRNRVRLIQFLMALTDEYEPVRASSLHQNPLPT 783
Query: 188 LDTCVGELLREEQRLLTQGTMSHDAFISEPVPVAYAAQSRGKGRDMRQVQCFTCKQFGHV 247
L+ + L EE RL + +A+A +
Sbjct: 784 LENALPCLKSEETRL---------QLVPPKADLAFAVTNN-------------------- 876
Query: 248 ARSCTAKFCKYCKQNGHVIFDCP 270
K C++C+++GH DCP
Sbjct: 877 ----ATKPCRHCQKSGHSFSDCP 933
>BF645022 weakly similar to GP|8777581|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (8%)
Length = 685
Score = 62.0 bits (149), Expect = 1e-09
Identities = 27/105 (25%), Positives = 54/105 (50%)
Frame = -2
Query: 14 VRFTGKNYPAWEFQFRMYVKGNKLWSHLDDVSKAPTEKAALEEWEYKDAQIISWILSSID 73
++ G NY W R + + + ++ PT LE+W+ + +I+W+L++I+
Sbjct: 330 IQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPKPTTPEKLEDWKAVQSMLIAWLLNTIE 151
Query: 74 PQMINNLRSFSTAQEMWNYLKRIYNQDNAAKRFQLELEIANYKQG 118
P + + L + A+ +W +LK+ + N A+ QL+ + KQG
Sbjct: 150 PSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKASLGECKQG 16
>AW691018 weakly similar to GP|2462935|em open reading frame 1 {Brassica
oleracea}, partial (12%)
Length = 639
Score = 57.0 bits (136), Expect = 5e-08
Identities = 29/116 (25%), Positives = 60/116 (51%), Gaps = 1/116 (0%)
Frame = +2
Query: 18 GKNYPAWEFQFRMYVKGNKLWSHLDDVSKAPT-EKAALEEWEYKDAQIISWILSSIDPQM 76
G NY W + + +D KAP+ + L W+ + +++WIL SI+P +
Sbjct: 215 GGNYGEWSRSMLLSLSAKNKLGLIDGTVKAPSADDPKLPLWKRCNDLVLTWILHSIEPDI 394
Query: 77 INNLRSFSTAQEMWNYLKRIYNQDNAAKRFQLELEIANYKQGNLYVQEFYSGFLNL 132
++ TA +W+ L ++Q + ++ +Q+ EI+ +QG+L + ++Y+ +L
Sbjct: 395 ARSVIFSDTAAAVWSDLHDRFSQGDESRIYQIRQEISECRQGSLLISDYYTKLKSL 562
>BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element polyprotein
[imported] - Arabidopsis thaliana, partial (2%)
Length = 608
Score = 53.5 bits (127), Expect = 5e-07
Identities = 27/124 (21%), Positives = 54/124 (42%), Gaps = 3/124 (2%)
Frame = +1
Query: 15 RFTGKNYPAWEFQFRMYVKGNKLWSHLDDVSKAPTE---KAALEEWEYKDAQIISWILSS 71
+ G NY +W + ++ K P+E A W ++ I+SW+ S
Sbjct: 172 KLNGTNYSSWSRSMVHALTAKNKVGFINGSIKTPSEVDQPAEYALWNRCNSMILSWLTHS 351
Query: 72 IDPQMINNLRSFSTAQEMWNYLKRIYNQDNAAKRFQLELEIANYKQGNLYVQEFYSGFLN 131
++P + + TA ++W K ++Q N +Q++ +A+ QG + +++
Sbjct: 352 VEPDLAKGVIHAKTACQVWEDFKDQFSQKNIPAIYQIQKSLASLSQGTVSASTYFTKIKG 531
Query: 132 LWTE 135
LW E
Sbjct: 532 LWDE 543
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 52.8 bits (125), Expect = 9e-07
Identities = 23/43 (53%), Positives = 31/43 (71%)
Frame = +1
Query: 614 NTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFWVEALSTVCF 656
+TPQQNG+AER NR LL+ TR++L A + FW EA+ T C+
Sbjct: 82 HTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACY 210
>BQ122291
Length = 487
Score = 52.4 bits (124), Expect = 1e-06
Identities = 45/179 (25%), Positives = 77/179 (42%), Gaps = 1/179 (0%)
Frame = -2
Query: 42 DDVSKAPTEKAALEEWEYKDAQIISWILSSIDPQMINNLRSFSTAQEMWNYLKRIYNQDN 101
D +A T A EWE KD+ + +WILS I P +++ + ++W+ +
Sbjct: 486 DAAREAGTRNPAYTEWEDKDSLLCTWILSRISPSLLSRFVLLRHSWQVWDEIHSYCFTQM 307
Query: 102 AAKRFQLELEIANYKQGNLYVQEFYSGFLNLWTEHSAIIHADVPKASLAAVQE-VYNTSR 160
+ QL E+ + +G+ V EF I SLA++ + V +
Sbjct: 306 KTRSRQLRSELRSITKGSRTVSEF-------------IARIRAISESLASIGDPVSHRDL 166
Query: 161 RDQFLMKLRPEFEVVRGALLNRNPVPSLDTCVGELLREEQRLLTQGTMSHDAFISEPVP 219
+ L L EF+ + ++ ++ V SLD +LL +E R A ++EPVP
Sbjct: 165 IEVVLEALPEEFDPIVASVNAKSEVVSLDELESQLLTQESR----KEKFKKAAMNEPVP 1
>TC93152
Length = 647
Score = 50.1 bits (118), Expect = 6e-06
Identities = 25/89 (28%), Positives = 43/89 (48%), Gaps = 2/89 (2%)
Frame = +3
Query: 17 TGKNYPAWEFQFRMYVKGNKLWSHLDDVSKA--PTEKAALEEWEYKDAQIISWILSSIDP 74
T NY W R Y+ G LW + +S++ P + E W ++ + + I + P
Sbjct: 153 TKDNYDYWSCLVRNYLLGQDLWGFVTSISESTGPRSRRETEVWNRRNGKALHIIQLACGP 332
Query: 75 QMINNLRSFSTAQEMWNYLKRIYNQDNAA 103
+ IN ++ +A+E WN L Y+ D +A
Sbjct: 333 ENINLIKDLQSAREAWNELSTHYSSDLSA 419
>AW773859
Length = 538
Score = 48.9 bits (115), Expect = 1e-05
Identities = 22/68 (32%), Positives = 34/68 (49%)
Frame = -3
Query: 460 WHRKLGHPNSTVLSHLFKTGLLGNKQVVCTASISCPVCKLAKSKTLPFPSGAHRASNCFE 519
WH +LGH ++ L L + + C +C ++ K LPF +RAS C+E
Sbjct: 227 WHFRLGHLSNRKLLSLHSNFPF----ITIDQNSVCDICHYSRHKKLPFQLSTNRASKCYE 60
Query: 520 MIHSDVWG 527
+ H D+WG
Sbjct: 59 LFHFDIWG 36
>AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (18%)
Length = 567
Score = 46.6 bits (109), Expect = 6e-05
Identities = 34/154 (22%), Positives = 71/154 (46%), Gaps = 7/154 (4%)
Frame = +3
Query: 331 SRPWFLDSGASNHMTGSSEYLHNLHSYHGNQQIQIADGNKLSITDVGDI-------NSDF 383
S+ W +DSG ++HMT + L+ ++++ +G + + +G +
Sbjct: 99 SKYWLIDSGCTHHMTHDRDLFKELNK-STISKVRMLNGAHIEVEGIGTVLVKSHSGYKQI 275
Query: 384 QDVLVSPGLASNLLSVGQLVDNNCNVNFSRAGCLVQEQVSGKVIAKGPKVGRLFPLQFIS 443
+VL +P L +LLSV QL+ V F C++++Q + K + + R F L +
Sbjct: 276 SNVLYAPKLNQSLLSVPQLLTKGYKVLFEHEKCVIKDQ-NNKEVLRAQMKARKFVLDLKN 452
Query: 444 SHLSLACNNVLNSYEDWHRKLGHPNSTVLSHLFK 477
+++ + ++++ K H ++ VL K
Sbjct: 453 GYIT----DEDEKKKEYNFKEDHNDACVLKKFIK 542
>AW684891
Length = 539
Score = 46.6 bits (109), Expect = 6e-05
Identities = 24/83 (28%), Positives = 43/83 (50%)
Frame = -3
Query: 541 VTFIDDYSRFTWIYFLRSKSEVFSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQEY 600
+T I Y ++ F+ + + + +F K + QF S++ SN G Y+ +Y
Sbjct: 369 LTLIQIYGXKWFLAFIHNSTSIT*LFLK-KHEIWFQFVKSIERLCSNKGKVYVDQNLSQY 193
Query: 601 LQHKGILSQRSCPNTPQQNGLAE 623
L+ G+ + C N+PQQNG++E
Sbjct: 192 LEENGVSQELKCVNSPQQNGVSE 124
>BF631997 weakly similar to GP|18542925|gb Putative pol polyprotein {Oryza
sativa}, partial (6%)
Length = 650
Score = 44.3 bits (103), Expect = 3e-04
Identities = 24/73 (32%), Positives = 38/73 (51%)
Frame = -2
Query: 554 YFLRSKSEVFSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQEYLQHKGILSQRSCP 613
+ ++ KSE F + + + +F+ N+G E S EF E + + I Q +
Sbjct: 643 FMMKHKSEAFYFSRNGRFWSRVKQGRR*NVFKLNNGLEICSAEFNELCKEEHITRQYTVR 464
Query: 614 NTPQQNGLAERKN 626
NTPQ+NG+AER N
Sbjct: 463 NTPQKNGVAERMN 425
>BI262818 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (24%)
Length = 615
Score = 43.9 bits (102), Expect = 4e-04
Identities = 31/135 (22%), Positives = 58/135 (42%), Gaps = 8/135 (5%)
Frame = +1
Query: 5 TSEKAKDFCVRFTGKNYPAWEFQFRMYVKGNKLWSHLDD------VSKAPTEKAALEEWE 58
TS KA V F G+NY W + Y++ N LW +++ +S PT E
Sbjct: 154 TSYKAPPLPV-FDGENYHIWAARIEAYLEANDLWEAVEEDYEVLPLSDNPTMAQIKNHKE 330
Query: 59 YK--DAQIISWILSSIDPQMINNLRSFSTAQEMWNYLKRIYNQDNAAKRFQLELEIANYK 116
K ++ + + +++ ++ + + +A E+WN+LK Y D + Q I ++
Sbjct: 331 RKTRKSKARATLFAAVSEEIFTRIMTIKSAFEIWNFLKTEYEGDERIRGMQALNLIREFE 510
Query: 117 QGNLYVQEFYSGFLN 131
+ E + N
Sbjct: 511 MQKMKESETIKEYAN 555
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 42.4 bits (98), Expect = 0.001
Identities = 20/49 (40%), Positives = 27/49 (54%), Gaps = 5/49 (10%)
Frame = +1
Query: 227 RGKGRD--MRQVQCFTCKQFGHVARSCTA---KFCKYCKQNGHVIFDCP 270
RG RD R V C +C+QFGH++R C C+ C GH ++CP
Sbjct: 874 RGGYRDGGFRDVVCRSCQQFGHMSRDCMGGPLMICQNCGGRGHQAYECP 1020
Score = 38.1 bits (87), Expect = 0.023
Identities = 16/37 (43%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Frame = +1
Query: 238 CFTCKQFGHVARSCT-AKFCKYCKQNGHVIFDCPIRP 273
C C + GH+A CT K C C++ GH+ DCP P
Sbjct: 673 CNNCYKQGHIAVECTNEKACNNCRKTGHLARDCPNDP 783
Score = 35.4 bits (80), Expect = 0.15
Identities = 14/34 (41%), Positives = 18/34 (52%), Gaps = 1/34 (2%)
Frame = +1
Query: 238 CFTCKQFGHVARSCTAK-FCKYCKQNGHVIFDCP 270
C C GH+A C+ K C CK+ GH+ CP
Sbjct: 481 CHNCSLPGHIASECSTKSLCWNCKEPGHMASSCP 582
Score = 34.3 bits (77), Expect = 0.33
Identities = 14/40 (35%), Positives = 19/40 (47%), Gaps = 8/40 (20%)
Frame = +1
Query: 238 CFTCKQFGHVARSCTA--------KFCKYCKQNGHVIFDC 269
C TC + GH AR CT + C C + GH+ +C
Sbjct: 595 CHTCGKAGHRARECTVPQKPPGDLRLCNNCYKQGHIAVEC 714
Score = 33.9 bits (76), Expect = 0.43
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 3/40 (7%)
Frame = +1
Query: 238 CFTCKQFGHVARSCTAK-FCKYCKQNGHVIFDC--PIRPP 274
C+ CK+ GH+A SC + C C + GH +C P +PP
Sbjct: 538 CWNCKEPGHMASSCPNEGICHTCGKAGHRARECTVPQKPP 657
Score = 30.0 bits (66), Expect = 6.3
Identities = 12/33 (36%), Positives = 15/33 (45%), Gaps = 1/33 (3%)
Frame = +1
Query: 238 CFTCKQFGHVARSC-TAKFCKYCKQNGHVIFDC 269
C CK+ GH R C C C GH+ +C
Sbjct: 424 CKNCKRPGHYVRECPNVAVCHNCSLPGHIASEC 522
>BI266628
Length = 536
Score = 40.0 bits (92), Expect = 0.006
Identities = 22/76 (28%), Positives = 41/76 (53%)
Frame = -1
Query: 42 DDVSKAPTEKAALEEWEYKDAQIISWILSSIDPQMINNLRSFSTAQEMWNYLKRIYNQDN 101
D+ +A T+K A EWE +D+ + +WILS+I P +++ F + W ++ I
Sbjct: 311 DEAREAGTKKPAYTEWEEQDSLLCTWILSTI*PSLLSR---FVLLRHSWFGMRFIVTASC 141
Query: 102 AAKRFQLELEIANYKQ 117
AK L + A++++
Sbjct: 140 KAKILALPIVSASHER 93
>TC92872
Length = 923
Score = 38.1 bits (87), Expect = 0.023
Identities = 27/135 (20%), Positives = 58/135 (42%), Gaps = 10/135 (7%)
Frame = +1
Query: 15 RFTGKNYPAWEFQFRMYVKGNKLWSHLDDVSKAPTE----------KAALEEWEYKDAQI 64
+ + KN+ AW+ Q + G ++ H+D + TE +W D I
Sbjct: 106 KLSRKNFRAWKRQVTTLLAGIEVMGHIDGTTPIQTETIINNGVSAPNPDYTKWFTLDQLI 285
Query: 65 ISWILSSIDPQMINNLRSFSTAQEMWNYLKRIYNQDNAAKRFQLELEIANYKQGNLYVQE 124
I+ +LSS+ + S+ TA+ +W ++ +N + + + +I +G+ + +
Sbjct: 286 INLLLSSMTEADSISFASYETARTLWVAIESQFNNTSRSHVMSVTNQIQRCTKGDKSITD 465
Query: 125 FYSGFLNLWTEHSAI 139
+ +L E + I
Sbjct: 466 YLFSVKSLADELAVI 510
>BG450673 homologue to GP|12055499|emb NADH dehydrogenase subunit 2 {Sphagnum
fallax}, partial (2%)
Length = 678
Score = 34.7 bits (78), Expect(2) = 0.080
Identities = 14/49 (28%), Positives = 25/49 (50%)
Frame = +3
Query: 43 DVSKAPTEKAALEEWEYKDAQIISWILSSIDPQMINNLRSFSTAQEMWN 91
D+ P W +A + WI++S+D +I+N FST + +W+
Sbjct: 258 DLPPPPQTDPTFRRWGTDNAIVKGWIINSMDSSLISNFIRFSTTKLVWD 404
Score = 20.4 bits (41), Expect(2) = 0.080
Identities = 7/28 (25%), Positives = 13/28 (46%)
Frame = +1
Query: 14 VRFTGKNYPAWEFQFRMYVKGNKLWSHL 41
++ G NYP W + +++ HL
Sbjct: 190 IKLDGSNYPLWSQAKTNWDTSMEIFRHL 273
>TC82733 similar to GP|10177404|dbj|BAB10535.
gene_id:K24M7.12~pir||S42136~similar to unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 710
Score = 35.8 bits (81), Expect = 0.11
Identities = 15/40 (37%), Positives = 20/40 (49%), Gaps = 7/40 (17%)
Frame = +3
Query: 238 CFTCKQFGHVARSCTA-------KFCKYCKQNGHVIFDCP 270
CF CK H+A+ CT K C C++ GH +CP
Sbjct: 282 CFICKGLDHIAKFCTQKAEWEKNKICLRCRRRGHRAQNCP 401
>BG645911
Length = 617
Score = 35.4 bits (80), Expect = 0.15
Identities = 19/67 (28%), Positives = 33/67 (48%)
Frame = +3
Query: 34 GNKLWSHLDDVSKAPTEKAALEEWEYKDAQIISWILSSIDPQMINNLRSFSTAQEMWNYL 93
G LW H+ + + A +W+ K+ Q + I S + +R+ TAQE WN L
Sbjct: 6 GKDLWDHIVEGNL----DAQHSDWKSKNGQALHAIQLSCGHYTLGQIRNCVTAQEAWNRL 173
Query: 94 KRIYNQD 100
K ++++
Sbjct: 174 KAAFSEN 194
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.338 0.147 0.468
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,452,783
Number of Sequences: 36976
Number of extensions: 776132
Number of successful extensions: 8020
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 4555
Number of HSP's successfully gapped in prelim test: 498
Number of HSP's that attempted gapping in prelim test: 3156
Number of HSP's gapped (non-prelim): 5528
length of query: 1343
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1235
effective length of database: 5,021,319
effective search space: 6201328965
effective search space used: 6201328965
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144731.14


