

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144730.8 - phase: 0
(840 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
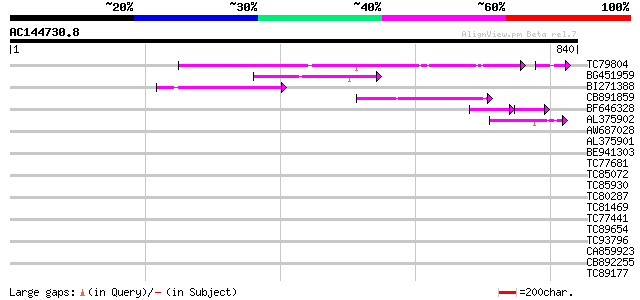
Score E
Sequences producing significant alignments: (bits) Value
TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like... 218 9e-63
BG451959 homologue to GP|6175165|gb| Mutator-like transposase {A... 109 5e-24
BI271388 similar to GP|10177186|db mutator-like transposase-like... 85 1e-16
CB891859 similar to GP|6175165|gb| Mutator-like transposase {Ara... 73 4e-13
BF646328 similar to PIR|F84533|F845 Mutator-like transposase [im... 59 4e-13
AL375902 similar to GP|9759134|dbj mutator-like transposase-like... 71 2e-12
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 41 0.002
AL375901 similar to GP|9759134|dbj mutator-like transposase-like... 40 0.005
BE941303 36 0.070
TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone d... 33 0.35
TC85072 similar to GP|18087573|gb|AAL58917.1 AT5g63950/MBM17_5 {... 33 0.46
TC85930 similar to GP|17065398|gb|AAL32853.1 Unknown protein {Ar... 32 1.0
TC80287 weakly similar to GP|12667516|gb|AAG61157.1 DBC2 {Homo s... 32 1.3
TC81469 similar to GP|21553812|gb|AAM62905.1 ripening-related pr... 31 2.3
TC77441 similar to GP|21592359|gb|AAM64310.1 unknown {Arabidopsi... 31 2.3
TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein... 31 2.3
TC93796 similar to PIR|H71402|H71402 probable kinesin - Arabidop... 31 2.3
CA859923 homologue to GP|4056491|gb|A unknown protein {Arabidops... 30 2.9
CB892255 30 2.9
TC89177 30 3.9
>TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like
transposase {Arabidopsis thaliana}, partial (57%)
Length = 2129
Score = 218 bits (556), Expect(2) = 9e-63
Identities = 143/521 (27%), Positives = 249/521 (47%), Gaps = 7/521 (1%)
Frame = +1
Query: 251 PQLNKDFKFKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECRDQ-CGYKVL 309
P +D F +G EF + F+ AI E + ++++++K++ R +C + C +++
Sbjct: 151 PDCMEDHNFVVGQEFPDVKAFRNAIKEAAIAQHFELRIIKSDLIRYFAKCASEGCPWRIR 330
Query: 310 CSKVGDRRTFQIKTLEGPHTCAPVLEN--KSANSRWVAKKAVPKMQVTKKMSVQEVFNEM 367
K+ + TF I++LEG HTC N A+ W+ +++ +++ +++
Sbjct: 331 AVKLPNASTFTIRSLEGTHTCGRNALNGHHQASVDWIVSFIEERLRDNINYKPKDILHDI 510
Query: 368 AVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSV 427
YG+ I +AWRA++ + I G +++ + ++ + E+K+ + K+ G
Sbjct: 511 HKQYGITIPYKQAWRAKERGLAAIYGSSEEGFYLLPSFCEEIKKTNPGSVAKVFTTGADS 690
Query: 428 TIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAF 487
Q F SFY G F++ C P V + G LK++Y L A D + FPLAF
Sbjct: 691 RFQRLFISFYASIHG----FVNGCLPIVALGGIQLKSKYLSTFLSATSFDADGGLFPLAF 858
Query: 488 GVVETECKETWKWFTQLLMEDIGED----KRYVFISDQQKGLLSVFDEMFDSIDHRLCLR 543
VV+ E E+W WF L + + + +F+SD QKG++ F H C+R
Sbjct: 859 AVVDVENDESWTWFLSELHNALEVNTECMPQIIFLSDGQKGIVDAIRRKFPRSSHAFCMR 1038
Query: 544 HLYANFKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCK 603
HL N K+F S++ L+ A AT + EKM E++++ A WL P +W
Sbjct: 1039HLSENIGKEF-KNSRLIHLLWSAAYATTINAFREKMAEIEEVSPNASMWLQHFHPSQWAL 1215
Query: 604 HAFTFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHN 663
F ++ L +NI E FN IL A++ PII + E I+ L K W
Sbjct: 1216VYFE-GTRYGHLSSNI-EEFNKWILEAQELPIIQVIERIQSKLKTEFDDRRLKSSSWCSV 1389
Query: 664 VMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGI 723
+ P +R+ + I A+ ++ S E F+V+ + IV+I SCSC W++ GI
Sbjct: 1390LTPSSERRMVEAINRASTYQVLKSDEVE-FEVISA--DRSDIVNIGSHSCSCRDWQLYGI 1560
Query: 724 PCRHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFAISPIN 764
PC H V+AL ++ + ++ + Y ++P++
Sbjct: 1561PCSHAVAALISSRKDVYAYTAKCFTVASYRDTVCRGVTPLS 1683
Score = 41.2 bits (95), Expect(2) = 9e-63
Identities = 19/52 (36%), Positives = 28/52 (53%)
Frame = +3
Query: 779 PPNYKNGPGRPRKLRIREFDENGSRMRRQGVAYRCTKCDQFGHNQRRCKSDV 830
PP ++ PGRP K RI D N R C++C+Q GH + CK+++
Sbjct: 1746 PPKFRRPPGRPEKKRICVEDHN-----RDKHTVHCSRCNQTGHYKTTCKAEM 1886
>BG451959 homologue to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (28%)
Length = 658
Score = 109 bits (272), Expect = 5e-24
Identities = 60/195 (30%), Positives = 102/195 (51%), Gaps = 4/195 (2%)
Frame = +2
Query: 361 QEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKI 420
+E+ E+ +G+ ++ +AWR ++ + + G ++ Y ++ +Y A +KR ++
Sbjct: 83 KEILEEIHRVHGITLSYKQAWRGKEHIMAAMRGSFEEGYRLLPQYCAHVKRT-NPGSIAS 259
Query: 421 NVAGPSVTIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPND 480
PS F + F G ++ACRP +G+D +LK++Y G LL+A G D +
Sbjct: 260 VYGNPSDNC---FQRLFISFQASIYGLLNACRPLLGLDRIYLKSKYLGTLLLATGFDGDG 430
Query: 481 QYFPLAFGVVETECKETWKWFT----QLLMEDIGEDKRYVFISDQQKGLLSVFDEMFDSI 536
FPLAFGVV+ E + W WF LL + R +SD+Q+G++ + F +
Sbjct: 431 ALFPLAFGVVDEENDDNWMWFLSKLHNLLEINTENMPRLTILSDRQQGIVDGVEANFPTA 610
Query: 537 DHRLCLRHLYANFKK 551
H C+RHL NF+K
Sbjct: 611 FHGFCMRHLSDNFRK 655
>BI271388 similar to GP|10177186|db mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (26%)
Length = 711
Score = 84.7 bits (208), Expect = 1e-16
Identities = 47/196 (23%), Positives = 101/196 (50%), Gaps = 3/196 (1%)
Frame = +2
Query: 218 DEEIDGDYLSEELGSSDPDDSNDDTIKYDQFRMPQLNKDFKFKIGMEFNTLVEFKEAIIE 277
+++++ D +E S +NDD + R Q ++ + FN+ EF+EA+ +
Sbjct: 20 EDQVEVDVANEAPAQSLCSGANDDKRQ----RAAQQWENTITGVDQRFNSFSEFREALHK 187
Query: 278 WNVLNGYQIKVLKNESYRVRVECRDQ-CGYKVLCSKVGDRRTFQIKTLEGPHTC--APVL 334
+++ +G+ + KN+S+RV V+C+ Q C +++ SK+ + IK + HTC + V
Sbjct: 188 YSIAHGFAYRYKKNDSHRVTVKCKSQGCPWRIYASKLSTTQLICIKKMTRDHTCEGSAVK 367
Query: 335 ENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGD 394
A WV K++ + +++ +++ YG+ + +AWRA++IA+ ++G
Sbjct: 368 AGYRATRGWVGNIIKEKLKASPNYRPKDIADDIKREYGIQLNYSQAWRAKEIAREQLQGS 547
Query: 395 ADKQYAMIRRYAAELK 410
+ Y + + ++K
Sbjct: 548 YKEAYTQLPFFCEKIK 595
>CB891859 similar to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (27%)
Length = 625
Score = 73.2 bits (178), Expect = 4e-13
Identities = 55/202 (27%), Positives = 89/202 (43%)
Frame = +1
Query: 514 RYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQ 573
R +SD+Q+G++ + F + H C+RHL +F+K+F + L A T +
Sbjct: 31 RLTILSDRQQGIVDGVEANFPTAFHGFCMRHLSDSFRKEFNNTMLVNLLWEAANCLTIIE 210
Query: 574 GWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQ 633
+ K+ E+++I A W+ V P+ W F + + L NI E+ N IL A
Sbjct: 211 -FEGKVMEIEEISQDAAYWIRRVPPRLWATAYFEGH-RFGHLTANIVEALNSWILEASGL 384
Query: 634 PIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQF 693
PII M E IR LM +W ++P + + + + A ++ + E
Sbjct: 385 PIIQMMECIRRQLMTWFNERRETSMQWTSILVPSAERSVAEALERARTYQVLRANEAEFE 564
Query: 694 QVMHTYNRQQFIVDIAKRSCSC 715
+ H IVDI R C C
Sbjct: 565 VISHEGTN---IVDIRNRCCLC 621
>BF646328 similar to PIR|F84533|F845 Mutator-like transposase [imported] -
Arabidopsis thaliana, partial (3%)
Length = 636
Score = 58.5 bits (140), Expect(2) = 4e-13
Identities = 24/66 (36%), Positives = 36/66 (54%)
Frame = -1
Query: 682 WKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEM 741
WK+TW G E + ++IV++A+R+C+C W + GIPC HV+ + E
Sbjct: 477 WKATWHGDMEMNNFNVSNETNKYIVNLAQRTCACRKWNLTGIPCAHVIPCIWHNGLADEN 298
Query: 742 FVDDYY 747
FV YY
Sbjct: 297 FVSSYY 280
Score = 34.7 bits (78), Expect(2) = 4e-13
Identities = 16/51 (31%), Positives = 25/51 (48%)
Frame = -3
Query: 749 RSKYALCYSFAISPINGMDMWPEVEAPELLPPNYKNGPGRPRKLRIREFDE 799
+S+ YS + P +G +WP + P + GRP+KLR + DE
Sbjct: 190 KSQCLATYSHIVLPSSGPKLWPVTNTEHINPLAKRRSAGRPKKLRKKANDE 38
>AL375902 similar to GP|9759134|dbj mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (20%)
Length = 542
Score = 70.9 bits (172), Expect = 2e-12
Identities = 41/127 (32%), Positives = 58/127 (45%), Gaps = 11/127 (8%)
Frame = +2
Query: 711 RSCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWP 770
R CSC W++ G PC H +AL MF + ++ Y + YS I PI W
Sbjct: 8 RECSCRRWQLYGYPCAHAAAALISCGHNAHMFAEPCFTVQSYRMAYSQMIYPIPDKSQWR 187
Query: 771 E-VEAPE----------LLPPNYKNGPGRPRKLRIREFDENGSRMRRQGVAYRCTKCDQF 819
E E E + PP + PGRP+K +R EN R +R +C +C
Sbjct: 188 EHGEGAEGGGGARVDIVIHPPKIRRPPGRPKKKVLRV--ENFKRPKR---VVQCGRCHML 352
Query: 820 GHNQRRC 826
GH+Q++C
Sbjct: 353 GHSQKKC 373
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 40.8 bits (94), Expect = 0.002
Identities = 31/148 (20%), Positives = 59/148 (38%), Gaps = 5/148 (3%)
Frame = +1
Query: 458 DGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKETWKWFTQLLMEDIGEDKRYVF 517
D + K +Y L I G + + Q ++E E E++KW +E +
Sbjct: 16 DTTYKKNKYNYPLCIFSGCNHHSQTIIFGVALLEDETIESYKWVLNRFLECMENKFPKAV 195
Query: 518 ISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQGWLE 577
++D + ++F HRLC HL+ N ++ + + G KA Y+ E
Sbjct: 196 VTDGDGSMREAIKQVFPDASHRLCAWHLHKNAQENI----KKTPFLEGFRKAMYSNFTPE 363
Query: 578 KMNE-----LKKIDLGAWEWLMAVEPKK 600
+ + ++K +L W++ K
Sbjct: 364 QFEDFWSELIQKNELEGNAWVIKTYANK 447
>AL375901 similar to GP|9759134|dbj mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (27%)
Length = 493
Score = 39.7 bits (91), Expect = 0.005
Identities = 29/144 (20%), Positives = 55/144 (38%)
Frame = +3
Query: 533 FDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEW 592
F S H CLR++ NF+ F +++ ++ A A + K+ E+ ++ W
Sbjct: 9 FPSASHGFCLRYVSENFRDTF-KNTKLVNIFWNAVYALTAAEFESKITEMIEVSQDVISW 185
Query: 593 LMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTT 652
P W F + ++E L + P++ M E+IR +
Sbjct: 186 FQHFPPFLWAVAYFDGVRYGHFTL-GVTELLYNWALECHELPVVQMMEYIRQQMTSWFND 362
Query: 653 SATKLQKWQHNVMPMPRKRLDKEI 676
+W ++P KR+ + I
Sbjct: 363 RREVGMEWTSILVPSAEKRISEAI 434
>BE941303
Length = 631
Score = 35.8 bits (81), Expect = 0.070
Identities = 20/53 (37%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Frame = -3
Query: 766 MDMWPEV-EAPELLPPNYKNGPGRPRKLRIREFDENGSRMRRQGVAYRCTKCD 817
MDMW +V + E++PP YK G RKL + ++ GVA+R CD
Sbjct: 617 MDMWLKVNDHEEIIPPLYKKGLSNLRKLELGNMMITD*GLKWPGVAHRRINCD 459
>TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone deacetylase
HD2-P39 {Glycine max}, partial (47%)
Length = 1233
Score = 33.5 bits (75), Expect = 0.35
Identities = 42/187 (22%), Positives = 78/187 (41%), Gaps = 16/187 (8%)
Frame = +2
Query: 77 HIAEEVSGYSVANKVDAHIFV------EHNVLDISVKVDVPSFFDLDVGYDEE-DVMVST 129
HI++ G + +K + + + + VL K +P LD+ ++E ++ ++
Sbjct: 146 HISQVALGEAKKDKPNESVVIYLKVGEQKLVLGTLTKDKIPQI-SLDIVLEKEFELSHNS 322
Query: 130 DDDSGKFVGF-------DDSEDESTTALEDGFEDVEVEAPANGNNRVTVNNKTLRIKKCG 182
S F G+ DDSE+E T ED ED+ + N + + L++ +
Sbjct: 323 KAASVHFCGYKAYYEDNDDSEEEGETDSED--EDIPL-ITTEQNGKPEPKAEELKVSEPK 493
Query: 183 SKTPKKKSPKKN--NSGRMNVIAPVSLLKNGKGKYVVDEEIDGDYLSEELGSSDPDDSND 240
K +P KN + + V+ P ++ D D +E+GSSD + N
Sbjct: 494 KADAKNAAPAKNAATAKHVKVVDPKD-----------EDSDDDDESDDEIGSSDDEMENA 640
Query: 241 DTIKYDQ 247
D+ D+
Sbjct: 641 DSDSEDE 661
Score = 31.2 bits (69), Expect = 1.7
Identities = 26/90 (28%), Positives = 40/90 (43%), Gaps = 2/90 (2%)
Frame = +2
Query: 106 VKVDVPSFFDLDVGYDEEDVMVSTDDDSGKFVGFDDSEDESTTALEDGFEDVEVEAPANG 165
VKV P D D + +D + S+DD+ + ED+S ED E+ V+ G
Sbjct: 548 VKVVDPKDEDSDDDDESDDEIGSSDDEMENADSDSEDEDDSD---EDDEEETPVKKVDQG 718
Query: 166 NNR--VTVNNKTLRIKKCGSKTPKKKSPKK 193
R + + + KK + TP+K KK
Sbjct: 719 KKRPNESASKTPISSKKSKNATPEKTDGKK 808
>TC85072 similar to GP|18087573|gb|AAL58917.1 AT5g63950/MBM17_5 {Arabidopsis
thaliana}, partial (1%)
Length = 724
Score = 33.1 bits (74), Expect = 0.46
Identities = 22/75 (29%), Positives = 34/75 (45%)
Frame = +3
Query: 169 VTVNNKTLRIKKCGSKTPKKKSPKKNNSGRMNVIAPVSLLKNGKGKYVVDEEIDGDYLSE 228
++ +NKT R +K+ KK P+ N + +S K + D+E+D
Sbjct: 162 ISFSNKTQRSSMAANKSVKK--PQSLNDSHYRFLQDLSAPPKPSSKSI-DDELDTPIQPR 332
Query: 229 ELGSSDPDDSNDDTI 243
L D DD +DDTI
Sbjct: 333 NLVYQDVDDDDDDTI 377
>TC85930 similar to GP|17065398|gb|AAL32853.1 Unknown protein {Arabidopsis
thaliana}, partial (62%)
Length = 1234
Score = 32.0 bits (71), Expect = 1.0
Identities = 26/91 (28%), Positives = 37/91 (40%), Gaps = 3/91 (3%)
Frame = -3
Query: 637 SMAEWIRHYLMRRMTTSATKLQKWQ---HNVMPMPRKRLDKEITLAAHWKSTWSGIGEQF 693
S+ WI H+ R +S + +W N + P R HW S + F
Sbjct: 818 SIHVWIMHHETR--ISSLLCILRWHIAIRNSISRPSNR---------HWSSWYRCKHFIF 672
Query: 694 QVMHTYNRQQFIVDIAKRSCSCNFWEIVGIP 724
QV+ +N Q I DI+ C CN + G P
Sbjct: 671 QVLPRHNVNQEIKDISSCDCCCNVILLKGSP 579
>TC80287 weakly similar to GP|12667516|gb|AAG61157.1 DBC2 {Homo sapiens},
partial (4%)
Length = 779
Score = 31.6 bits (70), Expect = 1.3
Identities = 33/101 (32%), Positives = 49/101 (47%), Gaps = 3/101 (2%)
Frame = -3
Query: 118 VGYDEEDVMVSTDDDSGKFVGFDDSEDESTTALEDGFEDVEVE-APANGNNRVTVNNKTL 176
VGY EE V DDD DD E E +DG +D++ E P + N++
Sbjct: 462 VGYSEEQVNHDDDDD-------DDDESE-----DDGDDDLDDELVPWDIGNKL----GRQ 331
Query: 177 RIKKCGSKTPKKKSPKKNNSGRMNVIAPVSLLKNG--KGKY 215
R++K G K+ S K NNS R +P+ ++ G +GK+
Sbjct: 330 RMRKLG----KRVSSKMNNSKR----SPILFVRPGCVRGKH 232
>TC81469 similar to GP|21553812|gb|AAM62905.1 ripening-related protein-like
{Arabidopsis thaliana}, partial (8%)
Length = 623
Score = 30.8 bits (68), Expect = 2.3
Identities = 22/77 (28%), Positives = 34/77 (43%)
Frame = +1
Query: 577 EKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPII 636
+ +NELK + +G +EW M+ + W AFT + C +N V + R
Sbjct: 109 QTLNELKHLRMGTFEWQMS-NAQTWASTAFTNGNSCINGLNRSDAEKKVKLEVKRKVTDA 285
Query: 637 SMAEWIRHYLMRRMTTS 653
SM YL+ R+ S
Sbjct: 286 SMFISNALYLINRLGES 336
>TC77441 similar to GP|21592359|gb|AAM64310.1 unknown {Arabidopsis
thaliana}, partial (98%)
Length = 1046
Score = 30.8 bits (68), Expect = 2.3
Identities = 21/67 (31%), Positives = 34/67 (50%)
Frame = +3
Query: 94 HIFVEHNVLDISVKVDVPSFFDLDVGYDEEDVMVSTDDDSGKFVGFDDSEDESTTALEDG 153
H+F+E DI + +P D D + DV V++DD+S DD +++ T AL
Sbjct: 354 HLFLEGTKRDIEDNI-IPRSVDAD----DSDVEVNSDDESD-----DDDDEDDTEALLAE 503
Query: 154 FEDVEVE 160
E ++ E
Sbjct: 504 LEQIKKE 524
>TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein F20D10.290 -
Arabidopsis thaliana, partial (65%)
Length = 1358
Score = 30.8 bits (68), Expect = 2.3
Identities = 11/21 (52%), Positives = 15/21 (71%)
Frame = +1
Query: 710 KRSCSCNFWEIVGIPCRHVVS 730
K SCSC +E GI CRH+++
Sbjct: 1144 KASCSCQMFEYSGIVCRHILA 1206
>TC93796 similar to PIR|H71402|H71402 probable kinesin - Arabidopsis
thaliana, partial (5%)
Length = 744
Score = 30.8 bits (68), Expect = 2.3
Identities = 10/21 (47%), Positives = 12/21 (56%)
Frame = +2
Query: 713 CSCNFWEIVGIPCRHVVSALG 733
C C +W +VGI C HV G
Sbjct: 647 CHCRYWRVVGINCLHVCGNFG 709
>CA859923 homologue to GP|4056491|gb|A unknown protein {Arabidopsis
thaliana}, partial (6%)
Length = 387
Score = 30.4 bits (67), Expect = 2.9
Identities = 15/39 (38%), Positives = 21/39 (53%)
Frame = +1
Query: 222 DGDYLSEELGSSDPDDSNDDTIKYDQFRMPQLNKDFKFK 260
D DY S+ +D ++ N + I+Y F P K FKFK
Sbjct: 67 DDDYFSDPDELADENELNANGIEYKDFFDPPKKKSFKFK 183
>CB892255
Length = 853
Score = 30.4 bits (67), Expect = 2.9
Identities = 13/36 (36%), Positives = 21/36 (58%)
Frame = +2
Query: 448 ISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYF 483
+ C P +G +GC + T+ GG+L+ V D +YF
Sbjct: 221 LKICYPILGANGCQI-TKNGGKLVCPVNTDAVKKYF 325
>TC89177
Length = 1201
Score = 30.0 bits (66), Expect = 3.9
Identities = 30/130 (23%), Positives = 60/130 (46%), Gaps = 13/130 (10%)
Frame = +2
Query: 80 EEVSGYSVANKVDAHIFVEHNVLDISVK-----VDVPSFFDLDVGYDEED-VMVSTDDDS 133
+ S +S A+ +D +++ +D + D P + DVG DE+D ++V+ +S
Sbjct: 659 DSASNHSGASNIDVGNSNDNDEVDAGYQSLLPTYDPPGYISDDVGVDEDDHILVNLGSES 838
Query: 134 GKFVGFDDSEDESTTALEDGFE-------DVEVEAPANGNNRVTVNNKTLRIKKCGSKTP 186
+G + S+ +++ + FE D + P ++ V V+ R+ GS++
Sbjct: 839 ---LGQNSSDKQNSLFSDPAFEKSNELFLDPTYDPPGYISDDVGVDEGDHRLVNLGSESL 1009
Query: 187 KKKSPKKNNS 196
+ S K NS
Sbjct: 1010GQISCFKQNS 1039
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,772,920
Number of Sequences: 36976
Number of extensions: 386252
Number of successful extensions: 1956
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 1926
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1941
length of query: 840
length of database: 9,014,727
effective HSP length: 104
effective length of query: 736
effective length of database: 5,169,223
effective search space: 3804548128
effective search space used: 3804548128
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144730.8


