

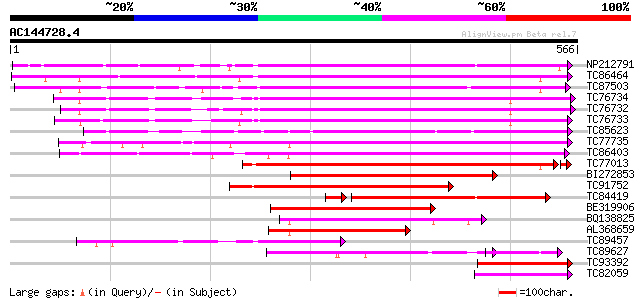
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144728.4 + phase: 0
(566 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1 364 e-101
TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Cit... 361 e-100
TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precurs... 357 5e-99
TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 352 2e-97
TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.... 345 3e-95
TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 336 2e-92
TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 ... 310 7e-85
TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectineste... 306 1e-83
TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF028... 304 5e-83
TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylestera... 291 1e-78
BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-... 261 4e-70
TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectineste... 217 1e-56
TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor ... 193 2e-52
BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precurso... 186 2e-47
BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like pr... 173 2e-43
AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886... 157 1e-38
TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE... 136 2e-32
TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pec... 114 1e-25
TC93392 similar to PIR|T49241|T49241 pectinesterase-like protein... 103 2e-22
TC82059 similar to GP|19424045|gb|AAL87311.1 unknown protein {Ar... 90 2e-18
>NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1
Length = 1698
Score = 364 bits (934), Expect = e-101
Identities = 213/574 (37%), Positives = 325/574 (56%), Gaps = 14/574 (2%)
Frame = +1
Query: 3 RKIFVSGVSLILVVGVALGVVALVRTNNGPADANNGGELTSHTKAVTAVCQNSDDHKFCA 62
+K + GVS IL+V + +GVVA+ T G D +++ K V +CQ++ + C
Sbjct: 40 KKHALLGVSCILLVAM-VGVVAVSLTKGG--DGEQKAHISNSQKNVDMLCQSTKFKETCH 210
Query: 63 DTLGSVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDLLEF 122
TL + S+ + IK + + E + K N + + + TK A++ C ++L++
Sbjct: 211 KTLEKASFSNMKNRIKGALGATEEELRKHIN--NSALYQELATDSMTKQAMEICNEVLDY 384
Query: 123 AIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGF---DTDGEKQVQSQ 179
A+D + S+ D H +++ A D+K WL ++QQ+CLDGF T + +
Sbjct: 385 AVDGIH-KSVGTLDQFDFHKLSEYAFDIKVWLTGTLSHQQTCLDGFVNTKTHAGETMAKV 561
Query: 180 LQTGSLDHVGKLTALALDVVTAITKVLAALDLDLNVKPS----SRRLFEVDEDGNPEWMS 235
L+T +L++ A+D++ ++++L PS SRRL + +DG P W+S
Sbjct: 562 LKTSM-----ELSSNAIDMMDVVSRILKGFH------PSQYGVSRRL--LSDDGIPSWVS 702
Query: 236 GADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEY 295
R LLA G +V NAVVA+DGSG+FKT+ DA+ + P + +VIYVKAGVY E
Sbjct: 703 DGHRHLLA----GGNVKANAVVAQDGSGQFKTLTDALKTVPPTNAAPFVIYVKAGVYKET 870
Query: 296 IQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGA 355
+ + K + + GDGPTKT TG N+ DG+ T +TATF F+AK + FENTAG
Sbjct: 871 VNVAKEMNYVTVIGDGPTKTKFTGSLNYADGINTYKTATFGVNGANFMAKDIGFENTAGT 1050
Query: 356 NKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQN 415
+K QAVALRV D++ F +C + G+QDTL+ + RQFYR+C ISGT+DF+FG A V QN
Sbjct: 1051SKFQAVALRVTADQAIFHNCQMDGFQDTLFVESQRQFYRDCAISGTIDFVFGDAFGVFQN 1230
Query: 416 SKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKA 475
K++ R P Q+ ++ A G ++N + +V + EPAL K+ S+L RPWK
Sbjct: 1231CKLICRVPAKGQKCLVTAGGRDKQNSASALVFLSSHFTGEPALTSVTPKL-SYLGRPWKL 1407
Query: 476 YSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVLSK 534
YS+ + M++TI + P+G++P G F DTC F EY N GPG++ RVKW G VL+
Sbjct: 1408YSKVVIMDSTIDAMFAPEGYMPMVGGAFKDTCTFYEYNNKGPGADTNLRVKWHGVKVLTS 1587
Query: 535 ADATKYTAAQWIE------GGVWLPATGIPFDLG 562
A +Y ++ E W+ +G+P+ LG
Sbjct: 1588NVAAEYYPGKFFEIVNATARDTWIVKSGVPYSLG 1689
>TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Citrus
sinensis}, partial (85%)
Length = 1993
Score = 361 bits (926), Expect = e-100
Identities = 216/578 (37%), Positives = 320/578 (54%), Gaps = 17/578 (2%)
Frame = +1
Query: 2 ERKIFVSGVSLILVVGVALGVVALVRTNNGPAD----ANNGGELTSHTKAVT-AVCQNSD 56
++K+ +S + +L+V + +VA N+ + A++ L+ H+ A+ + C +
Sbjct: 97 KKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTTL 276
Query: 57 DHKFCADTLGSV-----NTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKM 111
+ C + S ++ D I + + +V + +KL + + K+
Sbjct: 277 YPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLTKRE-KI 453
Query: 112 ALDDCKDLLEFAIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFD-T 170
AL DC + ++ +DEL+ + S + A DLK + + Q +CLDGF
Sbjct: 454 ALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSHD 633
Query: 171 DGEKQVQSQLQTGSLDHVGKLTALALDVVTAIT-KVLAALDLDLNVKPSSRRLFEVDED- 228
D +K+V+ LQ G + HV + + AL + +T K +A + V S++ ++E+
Sbjct: 634 DADKEVRKVLQEGQI-HVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEEN 810
Query: 229 --GNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIY 286
G PEW+S DR+LL G +V + VVA DGSG FKTV +A+ + P RYVI
Sbjct: 811 GVGWPEWISAGDRRLLQ----GSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIK 978
Query: 287 VKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKA 346
+KAGVY E +++ K K NI+ GDG T TIITG +N VDG T +AT + V F+A+
Sbjct: 979 IKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARD 1158
Query: 347 MAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIF 406
+ F+NTAG KHQAVALRV D SAF++C I YQDTLY H +RQF+ NC ISGTVDFIF
Sbjct: 1159ITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIF 1338
Query: 407 GYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVR 466
G ++ V QN I R+P + Q+N++ A G V N TG+V+Q C I L+ +
Sbjct: 1339GNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFP 1518
Query: 467 SFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVK 526
++L RPWK YSR +FM+++I D+I P G+ W G L+T + EY NTGPG+ RV
Sbjct: 1519TYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVT 1698
Query: 527 WG--KGVLSKADATKYTAAQWIEGGVWLPATGIPFDLG 562
W K + S A+A T +I G WL +TG PF LG
Sbjct: 1699WKGFKVITSAAEAQSSTPGNFIGGSSWLGSTGFPFSLG 1812
>TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precursor (EC
3.1.1.11) (Pectin methylesterase 3) (PE 3)., partial
(93%)
Length = 2088
Score = 357 bits (917), Expect = 5e-99
Identities = 210/569 (36%), Positives = 319/569 (55%), Gaps = 13/569 (2%)
Frame = +2
Query: 5 IFVSGVSLILVVGVALGVVALVRTNNGPADANNGGELTSHTKAVT--AVCQNSDDHKFCA 62
I +S + L+ V+ A+ + + + N + + N T T A + AVC+++ C
Sbjct: 152 IIISSIILVAVIIAAVAGILIHKHNTESSSSPNSLPNTELTPATSLKAVCESTQYPNSCF 331
Query: 63 DTLGSV---NTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQS-TKMALDDCKD 118
++ S+ NT+DP K +K +I+ + K L +EK + K A+ C +
Sbjct: 332 SSISSLPDSNTTDPEQLFKLSLKVAIDELSKL-----SLTRFSEKATEPRVKKAIGVCDN 496
Query: 119 LLEFAIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDTDGEKQVQS 178
+L ++D L S D + + + D++ WL A +CLD +V S
Sbjct: 497 VLADSLDRLNDSMSTIVDGGKMLSPA-KIRDVETWLSAALTDHDTCLDAVG-----EVNS 658
Query: 179 QLQTGSLDHVGKL----TALALDVVTAITKVLAALDLDLNVKPSSRRLFEVDEDGNPEWM 234
G + + ++ T A + + ++KV+ L + V RRL PEW+
Sbjct: 659 TAARGVIPEIERIMRNSTEFASNSLAIVSKVIRLLS-NFEVSNHHRRLL----GEFPEWL 823
Query: 235 SGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDE 294
A+R+LLA + P+AVVAKDGSG++KT+ +A+ K R+V+YVK GVY E
Sbjct: 824 GTAERRLLATVVN--ETVPDAVVAKDGSGQYKTIGEALKLVKKKSLQRFVVYVKKGVYVE 997
Query: 295 YIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAG 354
I +DK N++IYGDG T+T+++G +N++DG T +TATF+ +GFIAK + F NTAG
Sbjct: 998 NIDLDKNTWNVMIYGDGMTETVVSGSRNYIDGTPTFETATFAVKGKGFIAKDIQFLNTAG 1177
Query: 355 ANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQ 414
A+KHQAVA+R D+S F+ C+ GYQDTLYAH++RQFYR+C+I+GT+DFIFG A+ V Q
Sbjct: 1178ASKHQAVAMRSGSDQSVFYRCSFVGYQDTLYAHSNRQFYRDCDITGTIDFIFGNAAAVFQ 1357
Query: 415 NSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWK 474
N KI+ R+P +NQ N I A G N +G+V+Q P D L ++L RPWK
Sbjct: 1358NCKIMPRQPMSNQFNTITAQGKKDPNQNSGIVIQKSTFTTLPG---DNLIAPTYLGRPWK 1528
Query: 475 AYSRAIFMENTIGDLIQPDGFLPW-AGTQFLDTCFFAEYANTGPGSNVQARVKWG--KGV 531
+S I M++ IG ++P G++ W A + + +AEY NTGPG++V RVKW K
Sbjct: 1529DFSTTIIMKSEIGSFLKPVGWISWVANVEPPSSILYAEYQNTGPGADVPGRVKWAGYKPA 1708
Query: 532 LSKADATKYTAAQWIEGGVWLPATGIPFD 560
L DA K+T +I+G WLP+ + FD
Sbjct: 1709LGDEDAIKFTVDSFIQGPEWLPSASVQFD 1795
>TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, complete
Length = 1981
Score = 352 bits (903), Expect = 2e-97
Identities = 205/534 (38%), Positives = 294/534 (54%), Gaps = 12/534 (2%)
Frame = +3
Query: 44 HTKAVTAVCQNSDDHKFCADTLGSV-------NTSDPN-DYIKAVVKTSIESVIKAFNMT 95
H + VC+++ D C + V NT D + +++ S + KA +
Sbjct: 180 HFSSAPNVCEHAVDTNSCLTHVAEVVQGSTLDNTKDHKLSTLISLLTKSTTHIRKAMDTA 359
Query: 96 DKLAVENEKNNQSTKMALDDCKDLLEFAIDELQASSI-LAADNSSVHNVNDRAADLKNWL 154
+ ++ N+ + AL+ C+ L+ +++ + S + L DN D D WL
Sbjct: 360 N--VIKRRINSPREENALNVCEKLMNLSMERVWDSVLTLTKDNM------DSQQDAHTWL 515
Query: 155 GAVFAYQQSCLDGFDTDGEKQVQSQLQTGSLDHVGKLTALALDVVTAITKVLAALDLDLN 214
+V +CLDG + +++ +Q L A A + + VL D D
Sbjct: 516 SSVLTNHATCLDGLEGTSRAVMENDIQD--------LIARARSSLAVLVAVLPPKDHDEF 671
Query: 215 VKPSSRRLFEVDEDGNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINS 274
+ S F P W++ DR+LL + S G V N VVAKDGSGKFKTV +A+ S
Sbjct: 672 IDESLNGDF-------PSWVTSKDRRLL-ESSVG-DVKANVVVAKDGSGKFKTVAEAVAS 824
Query: 275 YPKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTAT 334
P RYVIYVK G+Y E ++I +K N+++ GDG TIITG N+VDG T QTAT
Sbjct: 825 APNKGTARYVIYVKKGIYKENVEIASSKTNVMLLGDGMDATIITGSLNYVDGTGTFQTAT 1004
Query: 335 FSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYR 394
+ V + FIA+ + F+NTAG KHQAVALRV D+S C I +QDTLYAH +RQFYR
Sbjct: 1005VAAVGDWFIAQDIGFQNTAGPQKHQAVALRVGSDRSVINRCKIDAFQDTLYAHTNRQFYR 1184
Query: 395 NCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMP 454
+ I+GT+DFIFG A+ V+Q K+V RKP ANQ N++ A G + N T +Q C+++P
Sbjct: 1185DSFITGTIDFIFGDAAVVLQKCKLVARKPMANQNNMVTAQGRIDPNQNTATSIQQCDVIP 1364
Query: 455 EPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPW--AGTQFLDTCFFAEY 512
L+P V+++L RPWK YSR + M++ +G I P G+ W A FL T ++ EY
Sbjct: 1365STDLKPVIGSVKTYLGRPWKKYSRTVVMQSLLGAHIDPTGWAEWDAASKDFLQTLYYGEY 1544
Query: 513 ANTGPGSNVQARVKW-GKGVLSKADATKYTAAQWIEGGVWLPATGIPFDLGFTK 565
N+GPG+ RVKW G +++ A+A K+T AQ I+G VWL TG+ F G K
Sbjct: 1545MNSGPGAGTSKRVKWPGYHIINTAEANKFTVAQLIQGNVWLKNTGVAFIAGL*K 1706
>TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11)
precursor - garden pea, complete
Length = 1860
Score = 345 bits (884), Expect = 3e-95
Identities = 200/530 (37%), Positives = 297/530 (55%), Gaps = 15/530 (2%)
Frame = +2
Query: 51 VCQNSDDHKFCADTLGSV-------NTSDPN-DYIKAVVKTSIESVIKAFNMTDKLAVEN 102
+C+++ D K C + V NT D + +++ S + KA + + ++
Sbjct: 194 LCEHALDTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTAN--VIKR 367
Query: 103 EKNNQSTKMALDDCKDLLEFAIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQ 162
N+ ++AL+DC++L++ ++D + S + N+ D D WL +V
Sbjct: 368 RVNSPREEIALNDCEELMDLSMDRVWDSVLTLTKNNI-----DSQHDAHTWLSSVLTNHA 532
Query: 163 SCLDGFDTDGEKQVQSQLQTGSLDHVGKLTALALDVVTAITKVLAALDLDLNVKPSSRRL 222
+CLDG + ++S L D+++ LA L ++V P
Sbjct: 533 TCLDGLEGSSRVVMESDLH---------------DLISRARSSLAVL---VSVLPPKAND 658
Query: 223 FEVDEDGN---PEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNH 279
+DE N P W++ DR+LL + S G + N VVA+DGSGKFKTV A+ S P N
Sbjct: 659 GFIDEKLNGDFPSWVTSKDRRLL-ESSVG-DIKANVVVAQDGSGKFKTVAQAVASAPDNG 832
Query: 280 QGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVA 339
+ RYVIYVK G Y E I+I K K N+++ GDG TIITG NF+DG T ++AT + V
Sbjct: 833 KTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVG 1012
Query: 340 EGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEIS 399
+GFIA+ + F+NTAG KHQAVALRV D+S C I +QDTLYAH++RQFYR+ I+
Sbjct: 1013DGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYIT 1192
Query: 400 GTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQ 459
GTVDFIFG A+ V Q SK+ RKP ANQ+N++ A G N T +Q C+++P L+
Sbjct: 1193GTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLK 1372
Query: 460 PDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPW--AGTQFLDTCFFAEYANTGP 517
P + ++++L RPWK YSR + +++ + I P G+ W A FL T ++ EY N+G
Sbjct: 1373PVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGA 1552
Query: 518 GSNVQARVKW-GKGVL-SKADATKYTAAQWIEGGVWLPATGIPFDLGFTK 565
G+ RV W G ++ + A+A+K+T Q I+G VWL TG+ F G K
Sbjct: 1553GAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL*K 1702
>TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, partial (98%)
Length = 2029
Score = 336 bits (861), Expect = 2e-92
Identities = 198/533 (37%), Positives = 292/533 (54%), Gaps = 15/533 (2%)
Frame = +2
Query: 45 TKAVTAVCQNSDDHKFCADTLGSVN-------TSDPNDYIKAVVKTSIESVIKAFNMTDK 97
T C+++ D K C + V+ T D N ++ + T + I+ N D
Sbjct: 191 TTVQNVYCEHAVDTKSCLAHVSEVSHVPTLVTTKDQNLHVLLSLLTKSTTHIQ--NAMDT 364
Query: 98 LAVENEK-NNQSTKMALDDCKDLLEFAIDELQASSILAADNSSVHNVNDRAADLKNWLGA 156
+V + N+ ++AL DC+ L++ +++ + + + N+ D D WL +
Sbjct: 365 ASVIKRRINSPREEIALSDCEQLMDLSMNRIWDTMLKLTKNNI-----DSQQDAHTWLSS 529
Query: 157 VFAYQQSCLDGFDTDGEKQVQSQLQTGSLDHVGKLTALALDVVTAITKVLAALDLDLNVK 216
V +CLDG + +++ LQ I++ ++L + L V
Sbjct: 530 VLTNHATCLDGLEGSSRVVMENDLQD------------------LISRARSSLAVFLVVF 655
Query: 217 PSSRRLFEVDED---GNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAIN 273
P R +DE P W++ DR+LL + + G + N VVA+DGSGKFKTV +A+
Sbjct: 656 PQKDRDQFIDETLIGEFPSWVTSKDRRLL-ETAVG-DIKANVVVAQDGSGKFKTVAEAVA 829
Query: 274 SYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTA 333
S P N + +YVIYVK G Y E ++I K N+++ GDG TIITG NF+DG T +++
Sbjct: 830 SAPDNGKTKYVIYVKKGTYKENVEIGSKKTNVMLVGDGMDATIITGNLNFIDGTTTFKSS 1009
Query: 334 TFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFY 393
T + V +GFIA+ + F+N AGA KHQAVALRV D+S C I +QDTLYAH++RQFY
Sbjct: 1010TVAAVGDGFIAQDIWFQNMAGAAKHQAVALRVGSDQSVINRCRIDAFQDTLYAHSNRQFY 1189
Query: 394 RNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIM 453
R+ I+GT+DFIFG A+ V Q K+V RKP ANQ N+ A G TG +Q C++
Sbjct: 1190RDSVITGTIDFIFGNAAVVFQKCKLVARKPMANQNNMFTAQGREDPGQNTGTSIQQCDLT 1369
Query: 454 PEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPW--AGTQFLDTCFFAE 511
P L+P +++FL RPWK YSR + M++ + I P G+ W A FL T ++ E
Sbjct: 1370PSSDLKPVVGSIKTFLGRPWKKYSRTVVMQSFLDSHIDPTGWAEWDAASKDFLQTLYYGE 1549
Query: 512 YANTGPGSNVQARVKW-GKGVL-SKADATKYTAAQWIEGGVWLPATGIPFDLG 562
Y N GPG+ RV W G V+ + A+A+K+T AQ I+G VWL TG+ F G
Sbjct: 1550YLNNGPGAGTAKRVTWPGYHVINTAAEASKFTVAQLIQGNVWLKNTGVAFTEG 1708
>TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 {Arabidopsis
thaliana}, partial (79%)
Length = 2442
Score = 310 bits (795), Expect = 7e-85
Identities = 187/491 (38%), Positives = 258/491 (52%), Gaps = 2/491 (0%)
Frame = +3
Query: 74 NDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDLLEFAIDELQASSIL 133
+D+ K ++ ++E K T AV + + K A DC +L +F + +L +
Sbjct: 741 SDFFKVSLEIAMERAKKGEENTH--AVGPKCRSPQEKAAWADCLELYDFTVQKLSQTKYT 914
Query: 134 AADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDTDGEKQVQSQLQTGSLDHVGKLTA 193
+ + WL ++C +GF G +T
Sbjct: 915 KCTQY----------ESQTWLSTALTNLETCKNGFYDLG------------------VTN 1010
Query: 194 LALDVVTAITKVLAALDLDLNVKPSSRRLFEVDEDGNPEWMSGADRKLLADMSTGMSVTP 253
L +++ L + L LN P + ++ DG P W+ DRKLL S
Sbjct: 1011 YVLPLLSNNVTKLLSNTLSLNKVPYQQPSYK---DGFPTWVKPGDRKLLQTSSAASKA-- 1175
Query: 254 NAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPT 313
N VVAKDGSGK+ TV A ++ P GRYVIYVKAGVY+E ++I KN+++ GDG
Sbjct: 1176 NVVVAKDGSGKYTTVKAATDAAPSG-SGRYVIYVKAGVYNEQVEIKA--KNVMLVGDGIG 1346
Query: 314 KTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFF 373
KTIITG K+ G T ++AT + +GFIA+ + F NTAGA HQAVA R D S F+
Sbjct: 1347 KTIITGSKSVGGGTTTFRSATVAATGDGFIAQDITFRNTAGAANHQAVAFRSGSDLSVFY 1526
Query: 374 DCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVA 433
C+ GYQDTLY H+ RQFYR C I GTVDFIFG A+ V+QN I R P A + + A
Sbjct: 1527 KCSFEGYQDTLYVHSERQFYRECNIYGTVDFIFGNAAVVLQNCNIFARNPPA-KTITVTA 1703
Query: 434 DGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPD 493
G N TG+++ N + + L P V+S+L RPW+ YSR +FM+ + I P
Sbjct: 1704 QGRTDPNQNTGIIIHNSRVSAQSDLNPS--SVKSYLGRPWQKYSRTVFMKTVLDGFINPA 1877
Query: 494 GFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVLSKA-DATKYTAAQWIEGGVW 551
G+LPW G LDT ++AEYANTG GS+ RV W G VL+ A A+ +T +I G W
Sbjct: 1878 GWLPWDGNFALDTLYYAEYANTGSGSSTSNRVTWKGYHVLTSASQASPFTVGNFIAGNSW 2057
Query: 552 LPATGIPFDLG 562
+ TG+PF G
Sbjct: 2058 IGNTGVPFTSG 2090
>TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectinesterase
{Arabidopsis thaliana}, partial (59%)
Length = 1963
Score = 306 bits (785), Expect = 1e-83
Identities = 192/545 (35%), Positives = 283/545 (51%), Gaps = 31/545 (5%)
Frame = +2
Query: 49 TAVCQNSDDHKFCADTLGSVNTS--DPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNN 106
+A C+ + K C L ++ +S DP +Y K SI+ +K +K+ ++ +
Sbjct: 146 SAACKTTLYPKLCRSMLSAIRSSPSDPYNY----GKFSIKQNLKVARKLEKVFIDFLNRH 313
Query: 107 QSTKM-------ALDDCKDLLEFAIDELQASS--ILAADNSSVHNVNDRAADLKNWLGAV 157
QS+ AL DCKDL +D L++ S + +A +SS + + ++++L AV
Sbjct: 314 QSSSSLNHEEVGALVDCKDLNSLNVDYLESISDELKSASSSSSSSDTELVDKIESYLSAV 493
Query: 158 FAYQQSCLDGFDTDGEKQVQSQLQTGSLDHVGKLTALALDVVT-AITKVLAALDLDLNVK 216
+C DG + + + L L + +++L +VT A++K + +
Sbjct: 494 ATNHYTCYDGLVVT-KSNIANALAV-PLKDATQFYSVSLGLVTEALSKNMKRNKTRKHGL 667
Query: 217 PSS---------------RRLFEVDEDGNPEWMSGADRKLLADMSTGMSVTPNAVVAKDG 261
P+ R + + + + +R L S G+ + +V+ G
Sbjct: 668 PNKSFKVRQPLEKLIKLLRTKYSCQKTSSNCTSTRTERILKESESHGILLNDFVLVSPYG 847
Query: 262 SGKFKTVLDAINSYPKN---HQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIIT 318
++ DAI + P N G Y+IYV+ G Y+EY+ + K K NIL+ GDG TIIT
Sbjct: 848 IANHTSIGDAIAAAPNNTKPEDGYYLIYVREGYYEEYVIVPKHKNNILLVGDGINNTIIT 1027
Query: 319 GKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIR 378
G + +DG T ++TF+ E FIA + F NTAG KHQAVA+R D S F+ C+
Sbjct: 1028GNHSVIDGWTTFNSSTFAVSGERFIAVDITFRNTAGPEKHQAVAVRNNADLSTFYRCSFE 1207
Query: 379 GYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQ 438
GYQDTLY H+ RQFYR+C+I GTVDFIFG A+ V QN I RKP NQ+N + A G
Sbjct: 1208GYQDTLYVHSVRQFYRDCKIYGTVDFIFGNAAVVFQNCNIYARKPLPNQKNAVTAQGRTD 1387
Query: 439 KNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPW 498
N TG+ +QNC I L D S+L RPWK YSR ++M++ IGD +QP G+L W
Sbjct: 1388PNQNTGISIQNCTIDAAQDLANDLNSTMSYLGRPWKIYSRTVYMQSYIGDFVQPSGWLEW 1567
Query: 499 AGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVLSKADATKYTAAQWIEGGVWLPATGI 557
GT LDT F+ E+ N GPGS RV+W G +L+ A +T + G WLP T I
Sbjct: 1568NGTVGLDTIFYGEFNNYGPGSVTNNRVQWPGHFLLNDTQAWNFTVLNFTLGNTWLPDTDI 1747
Query: 558 PFDLG 562
P+ G
Sbjct: 1748PYTEG 1762
>TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1929
Score = 304 bits (779), Expect = 5e-83
Identities = 192/534 (35%), Positives = 274/534 (50%), Gaps = 24/534 (4%)
Frame = +3
Query: 50 AVCQNSDDHKFCADTLGSVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQST 109
A C+++ D +C L N ++ DY + VK S+ K ++ +K + +
Sbjct: 132 AACKSTPDPTYCKSILPPQN-ANVYDYGRFSVKKSLSQSRKFSDLINKYLQRRSTLSTTA 308
Query: 110 KMALDDCKDLLEFAIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGF- 168
AL DC+ L + D L +SS + ++ + + +++ L A+ QQ+CLDG
Sbjct: 309 LRALQDCQSLSDLNFDFL-SSSFQTVNKTTKFLPSLQGENIQTLLSAILTNQQTCLDGLK 485
Query: 169 DTDGEKQVQSQLQTGSLDHVGKLTALALDVVTA-----------------ITKVLAALDL 211
DT ++ L T L + KL +++L T K L
Sbjct: 486 DTSSAWSFRNGL-TIPLSNDTKLYSVSLAFFTKGWVNPKTNKTSFPNSKHSNKGFKNGRL 662
Query: 212 DLNVKPSSRRLFEVDEDGNPEWMSGADRKLLADMSTGMSVTPNAVV--AKDGSGKFKTVL 269
L + +R ++E S + RKLL G V +V ++DGSG F T+
Sbjct: 663 PLKMTSKTRAIYE----------SVSRRKLLQSNQVGEDVVVRDIVTVSQDGSGNFTTIN 812
Query: 270 DAINSYPK---NHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDG 326
DAI + P + G ++IYV AGVY+EYI IDK K +++ GDG KTIITG + VDG
Sbjct: 813 DAIAAAPNKSVSSDGYFLIYVTAGVYEEYITIDKKKTYLMMIGDGINKTIITGNHSVVDG 992
Query: 327 VKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYA 386
T + TF+ V +GF+ M NTAGA KHQAVALR D S F+ C+ GYQDTLY
Sbjct: 993 WTTFGSPTFAVVGQGFVGVNMTIRNTAGAVKHQAVALRNGADLSTFYSCSFEGYQDTLYT 1172
Query: 387 HAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVV 446
H+ RQFYR C+I GTVDFIFG A V QN + R P + Q N I A G N TG
Sbjct: 1173HSLRQFYRECDIYGTVDFIFGNAKVVFQNCNLYPRLPMSGQFNAITAQGRTDPNQDTGTS 1352
Query: 447 LQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDT 506
+ NC I L V ++L RPWK YSR ++M+ + ++I G+ W G L T
Sbjct: 1353IHNCTIKATDDLAASNGAVSTYLGRPWKEYSRTVYMQTFMDNVINVAGWRAWDGEFALST 1532
Query: 507 CFFAEYANTGPGSNVQARVKW-GKGVLSKADATKYTAAQWIEGGVWLPATGIPF 559
++AE+ N+GPGS+ RV W G V++ DA +T A ++ G WLP TG+ +
Sbjct: 1533LYYAEFNNSGPGSSTDGRVTWQGYHVINATDAANFTVANFLLGDDWLPQTGVSY 1694
>TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylesterase isoform
alpha {Vigna radiata}, partial (94%)
Length = 1655
Score = 291 bits (745), Expect(2) = 1e-78
Identities = 145/319 (45%), Positives = 206/319 (64%), Gaps = 3/319 (0%)
Frame = +2
Query: 233 WMSGADRKLLADMSTGMSVTP-NAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGV 291
W+S DRKLL ++ T + VVAKDG+G F T+ +AI + P + R+VI++KAG
Sbjct: 41 WVSPKDRKLL---QAAVNQTKFDLVVAKDGTGNFATIGEAIAAAPNSSATRFVIHIKAGA 211
Query: 292 YDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFEN 351
Y E +++ K K N+++ GDG +T++ +N VDG T Q++TF+ V + FIAK + FEN
Sbjct: 212 YFENVEVIKKKTNLMLVGDGIGQTVVKASRNVVDGWTTFQSSTFAVVGDKFIAKGITFEN 391
Query: 352 TAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYAST 411
+AG +KHQAVA+R D SAF+ C+ YQDTLY H+ RQFYR C++ GTVDFIFG A+
Sbjct: 392 SAGPSKHQAVAVRNGADFSAFYQCSFVAYQDTLYVHSLRQFYRECDVYGTVDFIFGNAAV 571
Query: 412 VIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLAR 471
V QN + RKP+ Q+N+ A G N TG+ + NC+I L P + +S+L R
Sbjct: 572 VFQNCNLYARKPDPKQKNLFTAQGREDPNQNTGISILNCKIAAAADLIPVQSTFKSYLGR 751
Query: 472 PWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKWG--K 529
PWK YSR +++ + I +LI P G+L W GT LDT F+ EY N GPGSN ARV W K
Sbjct: 752 PWKKYSRTVYLNSFIDNLIDPPGWLEWNGTFALDTLFYGEYKNRGPGSNTSARVTWPGYK 931
Query: 530 GVLSKADATKYTAAQWIEG 548
+ + +A+++T Q+I+G
Sbjct: 932 VITNATEASQFTVRQFIQG 988
Score = 20.8 bits (42), Expect(2) = 1e-78
Identities = 7/11 (63%), Positives = 9/11 (81%)
Frame = +1
Query: 551 WLPATGIPFDL 561
WL +TG+PF L
Sbjct: 997 WLNSTGVPFFL 1029
>BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-like protein
- maize, partial (25%)
Length = 678
Score = 261 bits (668), Expect = 4e-70
Identities = 128/207 (61%), Positives = 152/207 (72%)
Frame = +1
Query: 281 GRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAE 340
GRY IYVKAGVYDEYI I K NIL+YGDGP KTI+TG+KN GVKT+QTATF+ A
Sbjct: 1 GRYTIYVKAGVYDEYITIPKDAVNILMYGDGPGKTIVTGRKNGAAGVKTMQTATFANTAL 180
Query: 341 GFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISG 400
GFI KAM FENTAG HQAVA R QGD SA C I GYQDTLY +RQFYRNC ISG
Sbjct: 181 GFIGKAMTFENTAGPAGHQAVAFRNQGDMSALVGCHILGYQDTLYVQTNRQFYRNCVISG 360
Query: 401 TVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQP 460
TVDFIFG ++T+IQ+S I+VR P NQ N I ADG+ + TG+V+Q C I+P+ AL P
Sbjct: 361 TVDFIFGTSATLIQDSTIIVRMPSPNQFNTITADGSYVNKLNTGIVIQGCNIVPKAALFP 540
Query: 461 DRLKVRSFLARPWKAYSRAIFMENTIG 487
R ++S+L RPWK ++ + ME+TIG
Sbjct: 541 QRFTIKSYLGRPWKVLTKTVVMESTIG 621
>TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectinesterase
{Arabidopsis thaliana}, partial (34%)
Length = 691
Score = 217 bits (552), Expect = 1e-56
Identities = 108/225 (48%), Positives = 144/225 (64%), Gaps = 1/225 (0%)
Frame = +3
Query: 220 RRLFEVD-EDGNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKN 278
RRL + ++ P W+ DRKLL + + + VVAKDGSGK+KT+ A+ P
Sbjct: 18 RRLLSLSHQNEEPNWLHHKDRKLL-QKDSDLKKKADIVVAKDGSGKYKTISAALKHVPNK 194
Query: 279 HQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTV 338
R VIYVK G+Y E ++++KTK N++I GDG TI++G NF+DG T TATF+
Sbjct: 195 SDKRTVIYVKKGIYYENVRVEKTKWNVMIIGDGMNVTIVSGSLNFIDGTPTFSTATFAVF 374
Query: 339 AEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEI 398
FIA+ + F+NTAG KHQAVAL D++ F+ C++ YQDTLYAH++RQFYR C I
Sbjct: 375 GRNFIARDIGFKNTAGPQKHQAVALMTSADQAVFYKCSMDAYQDTLYAHSNRQFYRECNI 554
Query: 399 SGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPT 443
GTVDFIFG ++ V+QN I+ R+P QQN I A G NM T
Sbjct: 555 YGTVDFIFGNSAVVLQNCNILPRQPMPGQQNTITAQGKTDPNMNT 689
>TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor (EC
3.1.1.11) (Pectin methylesterase) (PE) (P65). [Alfalfa],
partial (53%)
Length = 732
Score = 193 bits (490), Expect(2) = 2e-52
Identities = 91/200 (45%), Positives = 131/200 (65%), Gaps = 1/200 (0%)
Frame = +2
Query: 342 FIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGT 401
F A + FEN+AGA KHQAVALRV DK+ F++C + GYQDTLY + RQFYR+C I+GT
Sbjct: 86 FTALNVGFENSAGAAKHQAVALRVTADKALFYNCQMNGYQDTLYTQSKRQFYRDCTITGT 265
Query: 402 VDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPD 461
+DF+FG A V QN K++VRKP ANQQ ++ A G + + + +V QNC EP +
Sbjct: 266 IDFVFGDAVGVFQNCKLIVRKPMANQQCMVTAGGRTKVDSVSALVFQNCHFTGEPEVLTM 445
Query: 462 RLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNV 521
+ K+ ++L RPW+ +S+ + +++ I L P+G++PW G F +TC + EY N G G+
Sbjct: 446 QPKI-AYLGRPWRNFSKVVIVDSLIDGLFVPEGYMPWMGNLFKETCTYLEYNNKGAGAAT 622
Query: 522 QARVKW-GKGVLSKADATKY 540
+VKW G +S +A KY
Sbjct: 623 NLKVKWPGVKTISAGEAAKY 682
Score = 31.2 bits (69), Expect(2) = 2e-52
Identities = 13/21 (61%), Positives = 16/21 (75%)
Frame = +1
Query: 316 IITGKKNFVDGVKTIQTATFS 336
I TG K++ DGV+T TATFS
Sbjct: 7 IFTGSKSYADGVQTYNTATFS 69
>BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precursor (EC
3.1.1.11) (Pectin methylesterase) (PE). [Peach] {Prunus
persica}, partial (36%)
Length = 589
Score = 186 bits (472), Expect = 2e-47
Identities = 89/165 (53%), Positives = 120/165 (71%)
Frame = +2
Query: 261 GSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGK 320
GSG + V+DA+++ P++ RYVIYVK GVY E ++I K K NI++ G+G TII+G
Sbjct: 2 GSGDYAKVMDAVSAAPESSMKRYVIYVKKGVYVENVEIKKKKWNIMLIGEGMDATIISGS 181
Query: 321 KNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGY 380
+N+VDG T ++ATF+ GFIA+ ++F+NTAGA KHQAVALR D S F+ C I GY
Sbjct: 182 RNYVDGSTTFRSATFAVSGRGFIARDISFQNTAGAEKHQAVALRSDSDLSVFYRCGIFGY 361
Query: 381 QDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEA 425
QD+LY H RQFYR C+ISGTVDFIFG A+ V QN +I+ ++ +A
Sbjct: 362 QDSLYTHTMRQFYRECKISGTVDFIFGDATAVFQNCQILAKERDA 496
>BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like protein -
Arabidopsis thaliana, partial (36%)
Length = 658
Score = 173 bits (438), Expect = 2e-43
Identities = 95/218 (43%), Positives = 132/218 (59%), Gaps = 11/218 (5%)
Frame = +1
Query: 270 DAINSYPKN--HQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVD-G 326
+A+N+ P N + R+VIY+K GVY+E +++ K+N++ GDG KT+ITG N G
Sbjct: 4 EAVNAAPDNGVDRKRFVIYIKEGVYEETVRVPLKKRNVVFLGDGIGKTVITGSANVGQPG 183
Query: 327 VKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYA 386
+ T +AT + + +GF+AK + ENTAG + HQAVA R+ D S +C G QDTLYA
Sbjct: 184 MTTYNSATVAVLGDGFMAKDLTIENTAGPDAHQAVAFRLDSDLSVIENCEFLGNQDTLYA 363
Query: 387 HAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVR----KPEANQQNIIVADGTVQKNMP 442
H+ RQFY++C I G VDFIFG ++ + Q+ +I+VR KPE + N I A G
Sbjct: 364 HSLRQFYKSCRIVGNVDFIFGNSAAIFQDCQILVRPRQLKPEKGENNAITAHGRTDPAQS 543
Query: 443 TGVVLQNCEIMPEP---ALQPDRLKV-RSFLARPWKAY 476
TG V QNC I AL KV +++L RPWK Y
Sbjct: 544 TGFVFQNCLINGTEDYMALYHSNPKVHKNYLGRPWKEY 657
>AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (25%)
Length = 495
Score = 157 bits (397), Expect = 1e-38
Identities = 75/145 (51%), Positives = 100/145 (68%), Gaps = 3/145 (2%)
Frame = +1
Query: 259 KDGSGKFKTVLDAINSYPKN---HQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKT 315
+DGSG F T+ DAI + P N G ++I++ GVY+EY+ IDK KK +++ G+G +T
Sbjct: 61 QDGSGNFTTINDAIAAAPNNTVASDGYFLIFITEGVYEEYVSIDKKKKYLMMIGEGINQT 240
Query: 316 IITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDC 375
IITG +N DG T +ATF+ VA+GF+A + F NTAGA K+QAVALR D S F+ C
Sbjct: 241 IITGNRNVADGFTTFNSATFAVVAQGFVAVNITFRNTAGAAKNQAVALRSGADMSTFYSC 420
Query: 376 AIRGYQDTLYAHAHRQFYRNCEISG 400
+ GYQDTLY H+ RQFYR C+I G
Sbjct: 421 SFEGYQDTLYTHSLRQFYRECDIYG 495
>TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE8B
precursor (EC 3.1.1.11) (Pectin methylesterase) (PE).
[Peach] {Prunus persica}, partial (34%)
Length = 1415
Score = 136 bits (343), Expect = 2e-32
Identities = 95/280 (33%), Positives = 137/280 (48%), Gaps = 11/280 (3%)
Frame = +1
Query: 67 SVNTSDPNDYIKAVVKTSI-------ESVIKAFNMTDKLAVE--NEKNNQSTKMALDDCK 117
S S PND++ + ++ S E VI + N + T A+ DC
Sbjct: 163 STFASSPNDFVGSSLRVSTAKFANSAEEVITVLQKVISILSRFTNVFGHSRTSNAVSDCL 342
Query: 118 DLLEFAIDELQASSILAADNSSVHNVNDRA-ADLKNWLGAVFAYQQSCLDGFDTDGEKQV 176
DLL+ ++D+L S A N + DL+ WL AV Y +C++G E +
Sbjct: 343 DLLDMSLDQLNQSISAAQKPKEKDNSTGKLNCDLRTWLSAVLVYPDTCIEGL----EGSI 510
Query: 177 QSQLQTGSLDHVGKLTALAL-DVVTAITKVLAALDLDLNVKPSSRRLFEVDEDGNPEWMS 235
L + LDHV L A L +VV+ LA ++D P W+
Sbjct: 511 VKGLISSGLDHVMSLVANLLGEVVSGNDDQLAT-----------------NKDRFPSWIR 639
Query: 236 GADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEY 295
D KLL VT +AVVA DGSG + V+DA+++ P++ RYVIYVK GVY E
Sbjct: 640 DEDTKLLQ----ANGVTADAVVAADGSGDYAKVMDAVSAAPESSMKRYVIYVKKGVYVEN 807
Query: 296 IQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATF 335
++I K K NI++ G+G TII+G +N+VDG T ++ATF
Sbjct: 808 VEIKKKKWNIMLIGEGMDATIISGSRNYVDGSTTFRSATF 927
>TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pectin
methylesterase {Arabidopsis thaliana}, partial (33%)
Length = 1289
Score = 114 bits (284), Expect = 1e-25
Identities = 76/241 (31%), Positives = 119/241 (48%), Gaps = 10/241 (4%)
Frame = +3
Query: 257 VAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTI 316
V++DGS +FK++ +A+NS + R +I + G Y E I + KT I GD
Sbjct: 243 VSQDGSAQFKSITEALNSIQPYNIRRVIISIAPGYYREKIVVPKTLPFITFLGDVRDPPT 422
Query: 317 ITGKKNFV----DG--VKTIQTATFSTVAEGFIAKAMAFENTA----GANKHQAVALRVQ 366
ITG DG ++T +AT + A F+A + FENTA G+ QAVA+R+
Sbjct: 423 ITGNDTQSVTGSDGAQLRTFNSATVAVNASYFMAININFENTASFPIGSKVEQAVAVRIT 602
Query: 367 GDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEAN 426
G+K+AF++C G QDTLY H ++ NC I G+VDFI G+ ++ + I + AN
Sbjct: 603 GNKTAFYNCTFSGVQDTLYDHKGLHYFNNCTIKGSVDFICGHGKSLYEGCTI---RSIAN 773
Query: 427 QQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTI 486
I A + +G +N ++ + ++L RPW + F+ +
Sbjct: 774 NMTSITAQSGSNPSYDSGFSFKNSMVIGDGP---------TYLGRPWGKLFTSCFLLHLY 926
Query: 487 G 487
G
Sbjct: 927 G 929
Score = 53.5 bits (127), Expect = 2e-07
Identities = 24/77 (31%), Positives = 41/77 (53%)
Frame = +1
Query: 476 YSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKWGKGVLSKA 535
YS+ +F + + + P G+ W T+ ++ EY +GPGSN RV W + +L+
Sbjct: 895 YSQVVFSYTYMDNSVLPKGWEDWNDTKRYMNAYYGEYKCSGPGSNTAGRVPWAR-MLNDK 1071
Query: 536 DATKYTAAQWIEGGVWL 552
+A + Q+I+G WL
Sbjct: 1072EAQVFIGTQYIDGNTWL 1122
>TC93392 similar to PIR|T49241|T49241 pectinesterase-like protein -
Arabidopsis thaliana, partial (18%)
Length = 467
Score = 103 bits (257), Expect = 2e-22
Identities = 49/97 (50%), Positives = 70/97 (71%), Gaps = 2/97 (2%)
Frame = +2
Query: 468 FLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKW 527
+L RPWK YSR IFM++ + D I+P+G+L W G L+T ++AEY N+GPG+ V RVKW
Sbjct: 2 YLGRPWKTYSRTIFMQSYMSDAIRPEGWLEWNGNFALNTLYYAEYMNSGPGAGVANRVKW 181
Query: 528 -GKGVLS-KADATKYTAAQWIEGGVWLPATGIPFDLG 562
G VL+ ++ATK+T AQ+IEG + LP+TG+ + G
Sbjct: 182 SGYHVLNDSSEATKFTVAQFIEGNLGLPSTGVTYTSG 292
>TC82059 similar to GP|19424045|gb|AAL87311.1 unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 483
Score = 90.1 bits (222), Expect = 2e-18
Identities = 39/99 (39%), Positives = 59/99 (59%), Gaps = 1/99 (1%)
Frame = +1
Query: 465 VRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQAR 524
V+++L RPWK YSR ++M++ + I P G+ W G L T ++AEY N G GS+ R
Sbjct: 13 VKTYLGRPWKEYSRTVYMQSFMDSFINPSGWREWNGDFALSTLYYAEYDNRGAGSSTANR 192
Query: 525 VKW-GKGVLSKADATKYTAAQWIEGGVWLPATGIPFDLG 562
V W G V+ DA +T + ++ G W+P TG+P+ G
Sbjct: 193 VTWPGYHVIGATDAANFTVSNFVSGDDWIPQTGVPYMSG 309
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.133 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,447,058
Number of Sequences: 36976
Number of extensions: 170575
Number of successful extensions: 775
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 718
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 722
length of query: 566
length of database: 9,014,727
effective HSP length: 101
effective length of query: 465
effective length of database: 5,280,151
effective search space: 2455270215
effective search space used: 2455270215
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144728.4


