

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144728.10 + phase: 0 /pseudo
(1006 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
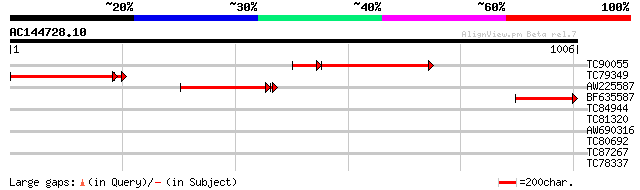
TC90055 homologue to PIR|S65668|S65668 preprotein translocase se... 388 e-131
TC79349 similar to PIR|S65668|S65668 preprotein translocase secA... 365 e-107
AW225587 homologue to PIR|S65668|S6 preprotein translocase secA ... 308 6e-85
BF635587 homologue to PIR|S65668|S6 preprotein translocase secA ... 217 2e-56
TC84944 similar to GP|10177507|dbj|BAB10901. serine protease-lik... 38 0.022
TC81320 similar to GP|13430618|gb|AAK25931.1 unknown protein {Ar... 32 1.2
AW690316 32 1.2
TC80692 similar to GP|20260382|gb|AAM13089.1 unknown protein {Ar... 30 3.6
TC87267 similar to PIR|T06242|T06242 aspartate kinase (EC 2.7.2.... 30 4.6
TC78337 similar to GP|9759359|dbj|BAB10014.1 GTP-binding protein... 30 6.1
>TC90055 homologue to PIR|S65668|S65668 preprotein translocase secA
precursor - garden pea, partial (24%)
Length = 750
Score = 388 bits (996), Expect(2) = e-131
Identities = 197/198 (99%), Positives = 197/198 (99%)
Frame = +2
Query: 554 RI*SLAVMLNLWHG*SFVRY*CQELLS*LKEILYR*RNLLLLRHGR*MTSCFHANYQIKI 613
RI*SLAVMLNLWHG*SFVRY*CQELLS*LKEILYR*RNLLLLRHGR*MTSCFHANYQIKI
Sbjct: 155 RI*SLAVMLNLWHG*SFVRY*CQELLS*LKEILYR*RNLLLLRHGR*MTSCFHANYQIKI 334
Query: 614 LSWLRRPCNWLLKHGERDH*LSWKQRNAYHILVKRAQPKMKSLPR*EMHFWKSARNIKSS 673
LSWLRRPCNWLLKHGERDH*LSWKQRNAYHILVKRAQPKMKSLPR*EMHFWKSARNIKSS
Sbjct: 335 LSWLRRPCNWLLKHGERDH*LSWKQRNAYHILVKRAQPKMKSLPR*EMHFWKSARNIKSS 514
Query: 674 PRKRGRRSWRLVD*L*WVPSVMNHGELTISCVVEVADKEIQEAHASF*VLKIIFFGYLAE 733
PRKRGRRSWRLVD*L*WVPSVMNHGELTISCVVEVADKEIQEAHASF*VLKIIFFGYLAE
Sbjct: 515 PRKRGRRSWRLVD*L*WVPSVMNHGELTISCVVEVADKEIQEAHASF*VLKIIFFGYLAE 694
Query: 734 TEFRA**KLSEWKTYLLS 751
TEF A**KLSEWKTYLLS
Sbjct: 695 TEFXA**KLSEWKTYLLS 748
Score = 99.4 bits (246), Expect(2) = e-131
Identities = 51/51 (100%), Positives = 51/51 (100%)
Frame = +1
Query: 503 VILCLSN*RKLESRTRFLMQNQKMLRGKQKL*HRVVVLGL*QLPPIWLVVG 553
VILCLSN*RKLESRTRFLMQNQKMLRGKQKL*HRVVVLGL*QLPPIWLVVG
Sbjct: 1 VILCLSN*RKLESRTRFLMQNQKMLRGKQKL*HRVVVLGL*QLPPIWLVVG 153
>TC79349 similar to PIR|S65668|S65668 preprotein translocase secA precursor
- garden pea, partial (19%)
Length = 857
Score = 365 bits (937), Expect(2) = e-107
Identities = 190/190 (100%), Positives = 190/190 (100%)
Frame = +3
Query: 1 MAASSLCSSFTSTTSHSQHRLHHRKTLNLPGSSFLSREFHLNSASVSKTRRSRSRRSGSV 60
MAASSLCSSFTSTTSHSQHRLHHRKTLNLPGSSFLSREFHLNSASVSKTRRSRSRRSGSV
Sbjct: 234 MAASSLCSSFTSTTSHSQHRLHHRKTLNLPGSSFLSREFHLNSASVSKTRRSRSRRSGSV 413
Query: 61 AVASLGGLLGGIFKGNDTGEATRKQYAATVNVINGLEANISKLSDSELRDKTFELRERAQ 120
AVASLGGLLGGIFKGNDTGEATRKQYAATVNVINGLEANISKLSDSELRDKTFELRERAQ
Sbjct: 414 AVASLGGLLGGIFKGNDTGEATRKQYAATVNVINGLEANISKLSDSELRDKTFELRERAQ 593
Query: 121 KRESLDSLLPEAFAVVREASKRVLGLRPFDVQLIGGMVLHKGEIAEMRTGEGKTLVAILP 180
KRESLDSLLPEAFAVVREASKRVLGLRPFDVQLIGGMVLHKGEIAEMRTGEGKTLVAILP
Sbjct: 594 KRESLDSLLPEAFAVVREASKRVLGLRPFDVQLIGGMVLHKGEIAEMRTGEGKTLVAILP 773
Query: 181 AYLNALVGKG 190
AYLNALVGKG
Sbjct: 774 AYLNALVGKG 803
Score = 43.1 bits (100), Expect(2) = e-107
Identities = 18/21 (85%), Positives = 19/21 (89%)
Frame = +1
Query: 187 VGKGVHVVTVNDYLARRDCEW 207
+GK VHVVTVNDYLA RDCEW
Sbjct: 793 LGKEVHVVTVNDYLAPRDCEW 855
>AW225587 homologue to PIR|S65668|S6 preprotein translocase secA precursor -
garden pea, partial (17%)
Length = 520
Score = 308 bits (788), Expect(2) = 6e-85
Identities = 160/160 (100%), Positives = 160/160 (100%)
Frame = +3
Query: 303 QKPLNVIYITLWMKNKNRF*FLNRATKMLKKF*LSKIYMIHENNGLHLS*MQLKQKNFFF 362
QKPLNVIYITLWMKNKNRF*FLNRATKMLKKF*LSKIYMIHENNGLHLS*MQLKQKNFFF
Sbjct: 3 QKPLNVIYITLWMKNKNRF*FLNRATKMLKKF*LSKIYMIHENNGLHLS*MQLKQKNFFF 182
Query: 363 EM*TT*FAEKRC*LLMSLLVE*CRGEGGVMDFIKQLKQRKGCPSKMKL*HWLLLVTRIFF 422
EM*TT*FAEKRC*LLMSLLVE*CRGEGGVMDFIKQLKQRKGCPSKMKL*HWLLLVTRIFF
Sbjct: 183 EM*TT*FAEKRC*LLMSLLVE*CRGEGGVMDFIKQLKQRKGCPSKMKL*HWLLLVTRIFF 362
Query: 423 CSSQNYAA*LALHQQKSQNLKAFTSLKLQSCLQTNL**ER 462
CSSQNYAA*LALHQQKSQNLKAFTSLKLQSCLQTNL**ER
Sbjct: 363 CSSQNYAA*LALHQQKSQNLKAFTSLKLQSCLQTNL**ER 482
Score = 26.2 bits (56), Expect(2) = 6e-85
Identities = 12/12 (100%), Positives = 12/12 (100%)
Frame = +1
Query: 463 MNLM*FSGLLEG 474
MNLM*FSGLLEG
Sbjct: 484 MNLM*FSGLLEG 519
>BF635587 homologue to PIR|S65668|S6 preprotein translocase secA precursor -
garden pea, partial (10%)
Length = 560
Score = 217 bits (553), Expect = 2e-56
Identities = 109/109 (100%), Positives = 109/109 (100%)
Frame = +1
Query: 898 SGS*S*AISIGCGKNICKH*NLFSKLWVYEGMRNGIHLLNINLRVTTFSWR*WHK*EEML 957
SGS*S*AISIGCGKNICKH*NLFSKLWVYEGMRNGIHLLNINLRVTTFSWR*WHK*EEML
Sbjct: 1 SGS*S*AISIGCGKNICKH*NLFSKLWVYEGMRNGIHLLNINLRVTTFSWR*WHK*EEML 180
Query: 958 YILYISLNPCC*SKIRIKLRIRNQVNGMHALGTIQIQIQLVQLSHQQVL 1006
YILYISLNPCC*SKIRIKLRIRNQVNGMHALGTIQIQIQLVQLSHQQVL
Sbjct: 181 YILYISLNPCC*SKIRIKLRIRNQVNGMHALGTIQIQIQLVQLSHQQVL 327
>TC84944 similar to GP|10177507|dbj|BAB10901. serine protease-like protein
{Arabidopsis thaliana}, partial (27%)
Length = 960
Score = 37.7 bits (86), Expect = 0.022
Identities = 29/92 (31%), Positives = 39/92 (41%), Gaps = 7/92 (7%)
Frame = -1
Query: 10 FTSTTSHSQHRLHHRKTLNLPGSSFLSREFHLNSASVSKTRRSRSRRSGSVAV-----AS 64
F T H HR+H R+ P L ++ S R R RR G+VAV A+
Sbjct: 534 FCMNTEHFHHRIHRRQH---PRRQSLPYRVRIHWCSTPSFNRRRDRRDGTVAVRAGKAAA 364
Query: 65 LGGLLG--GIFKGNDTGEATRKQYAATVNVIN 94
G LLG G+ G+ T + A V+ N
Sbjct: 363 CGWLLGNSGVLLGSATTATAELRRGAVVDEFN 268
>TC81320 similar to GP|13430618|gb|AAK25931.1 unknown protein {Arabidopsis
thaliana}, partial (6%)
Length = 635
Score = 32.0 bits (71), Expect = 1.2
Identities = 20/52 (38%), Positives = 29/52 (55%), Gaps = 1/52 (1%)
Frame = +1
Query: 6 LCS-SFTSTTSHSQHRLHHRKTLNLPGSSFLSREFHLNSASVSKTRRSRSRR 56
LCS +F S +HS+ H RK+ N G+S S F L S S S+ +R + +
Sbjct: 1 LCSHTFQSIRTHSKRTKHRRKSRNKQGASLSS--FQLLSTSASQKQRRKETK 150
>AW690316
Length = 606
Score = 32.0 bits (71), Expect = 1.2
Identities = 24/80 (30%), Positives = 42/80 (52%)
Frame = -1
Query: 33 SFLSREFHLNSASVSKTRRSRSRRSGSVAVASLGGLLGGIFKGNDTGEATRKQYAATVNV 92
SF SR ++ ++ ++ + R + GS A+LGG G IF ++ A + Q + T+ V
Sbjct: 276 SFTSR--NVRNSMITSSGRKITTADGS---AALGGG*GRIFITTNSTTAVQLQESVTLKV 112
Query: 93 INGLEANISKLSDSELRDKT 112
IN E + + +R+KT
Sbjct: 111 INKFELELEGEMKNGIREKT 52
>TC80692 similar to GP|20260382|gb|AAM13089.1 unknown protein {Arabidopsis
thaliana}, partial (25%)
Length = 1388
Score = 30.4 bits (67), Expect = 3.6
Identities = 18/55 (32%), Positives = 25/55 (44%), Gaps = 12/55 (21%)
Frame = -3
Query: 11 TSTTSHSQHRLHHRKT------------LNLPGSSFLSREFHLNSASVSKTRRSR 53
T+TT+ S H +HH KT N P S FL +FH + + K + R
Sbjct: 435 TTTTNRSNHLIHHPKTDTTHTLIFNFNSQNFPFSQFLQFQFHSHHS*HHKLNKKR 271
>TC87267 similar to PIR|T06242|T06242 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean, partial (24%)
Length = 817
Score = 30.0 bits (66), Expect = 4.6
Identities = 42/162 (25%), Positives = 65/162 (39%), Gaps = 26/162 (16%)
Frame = +3
Query: 3 ASSLCSSFTSTTSHSQHRLHHRKTLNLPGSS-----FLSREFHLNSASVSKTRRSRSRRS 57
ASSL SS + + S L H +P S LSR FH ++ RR S S
Sbjct: 54 ASSLSSSLSHFSRISVTSLQHDYNNKIPADSQCRHFLLSRRFHSLRKGITLPRRRESPSS 233
Query: 58 G----------SVAVASLGGLLGGI-----FKGNDTGEATRKQYAATVNVINGLEANISK 102
G +VAV G F G G + R + + + + E +
Sbjct: 234 GICASLTDVSVNVAVEEKELSKGDSWSVHKFGGTCMGSSQRIKNVGDIVLNDDSERKLVV 413
Query: 103 LSD-SELRDKTFELRERAQKRE-----SLDSLLPEAFAVVRE 138
+S S++ D ++L +AQ R+ SLD++L + A +
Sbjct: 414 VSAMSKVTDMMYDLINKAQSRDESYISSLDAVLEKHSATAHD 539
>TC78337 similar to GP|9759359|dbj|BAB10014.1 GTP-binding protein LepA
homolog {Arabidopsis thaliana}, partial (40%)
Length = 1035
Score = 29.6 bits (65), Expect = 6.1
Identities = 34/131 (25%), Positives = 57/131 (42%), Gaps = 5/131 (3%)
Frame = +1
Query: 3 ASSLCSSFTSTTSHSQHRLHHRKTLNLPGSSFLSREFHLNSASVSKTRRSRSRRSGSV-- 60
A+ LC + +Q LHH + + +SF S S SK R + SRR+ SV
Sbjct: 16 ATELCGGSLFLSVKTQRLLHHSQRTPITATSFFS--------SSSKPRFTFSRRNSSVFC 171
Query: 61 AVASLGGLLGGIFKGNDTGEATRKQYAATV---NVINGLEANISKLSDSELRDKTFELRE 117
VAS + D + K + + ++I ++ S L+D +L T + +
Sbjct: 172 RVASTDTVSNFDSPAKDGQDRLSKVPVSNIRNFSIIAHIDHGKSTLAD-KLLQVTGTVPQ 348
Query: 118 RAQKRESLDSL 128
R K + LD++
Sbjct: 349 REMKEQFLDNM 381
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.353 0.155 0.518
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,016,053
Number of Sequences: 36976
Number of extensions: 450309
Number of successful extensions: 5010
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 1533
Number of HSP's successfully gapped in prelim test: 232
Number of HSP's that attempted gapping in prelim test: 3416
Number of HSP's gapped (non-prelim): 1908
length of query: 1006
length of database: 9,014,727
effective HSP length: 106
effective length of query: 900
effective length of database: 5,095,271
effective search space: 4585743900
effective search space used: 4585743900
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.5 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144728.10


