

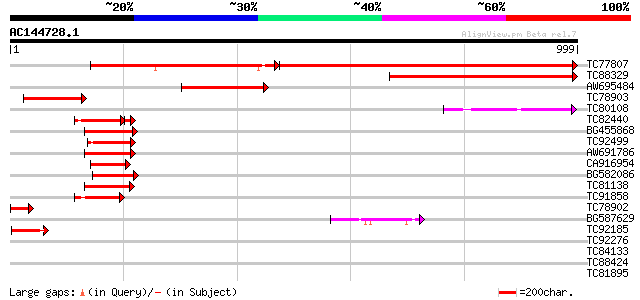
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144728.1 - phase: 0
(999 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77807 similar to PIR|T52601|T52601 squamosa promoter binding p... 1064 0.0
TC88329 similar to PIR|T47827|T47827 squamosa promoter binding p... 533 e-151
AW695484 similar to PIR|T52602|T526 squamosa promoter binding pr... 228 7e-60
TC78903 225 7e-59
TC80108 similar to GP|22045251|gb|AAL69483.2 putative SPL1-relat... 138 1e-32
TC82440 similar to PIR|H96793|H96793 unknown protein F14G6.18 [i... 109 4e-30
BG455868 similar to SP|Q38741|SBP1_ Squamosa-promoter binding pr... 123 4e-28
TC92499 homologue to PIR|T52594|T52594 squamosa promoter binding... 122 5e-28
AW691786 similar to PIR|T52298|T52 squamosa promoter binding pro... 116 5e-26
CA916954 similar to PIR|T52592|T525 squamosa-promoter binding pr... 103 4e-22
BG582086 similar to PIR|T52603|T526 squamosa promoter binding pr... 102 7e-22
TC81138 similar to PIR|T52606|T52606 squamosa promoter binding p... 97 2e-20
TC91858 similar to PIR|T52592|T52592 squamosa-promoter binding p... 92 1e-18
TC78902 89 8e-18
BG587629 similar to PIR|T52605|T526 squamosa promoter binding pr... 65 1e-10
TC92185 similar to GP|13374857|emb|CAC34491. putative protein {A... 56 6e-08
TC92276 similar to GP|15529228|gb|AAK97708.1 At1g03950/F21M11_12... 34 0.32
TC84133 weakly similar to GP|22093574|dbj|BAC06871. contains EST... 33 0.71
TC88424 similar to GP|22136724|gb|AAM91681.1 unknown protein {Ar... 32 0.93
TC81895 weakly similar to PIR|T48577|T48577 hypothetical protein... 32 1.2
>TC77807 similar to PIR|T52601|T52601 squamosa promoter binding protein 1
[imported] - Arabidopsis thaliana, partial (55%)
Length = 2923
Score = 1064 bits (2751), Expect(2) = 0.0
Identities = 524/524 (100%), Positives = 524/524 (100%)
Frame = +1
Query: 476 QSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGWVHIRV 535
QSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGWVHIRV
Sbjct: 1021 QSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGWVHIRV 1200
Query: 536 QHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPATRLMC 595
QHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPATRLMC
Sbjct: 1201 QHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPATRLMC 1380
Query: 596 ALEGKYLVCEDAHESTDQYSEELDELQCIQFSCSVPVSNGRGFIEIEDQGLSSSFFPFIV 655
ALEGKYLVCEDAHESTDQYSEELDELQCIQFSCSVPVSNGRGFIEIEDQGLSSSFFPFIV
Sbjct: 1381 ALEGKYLVCEDAHESTDQYSEELDELQCIQFSCSVPVSNGRGFIEIEDQGLSSSFFPFIV 1560
Query: 656 AEEDVCTEIRVLEPLLESSETDPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQLKYRMVNL 715
AEEDVCTEIRVLEPLLESSETDPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQLKYRMVNL
Sbjct: 1561 AEEDVCTEIRVLEPLLESSETDPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQLKYRMVNL 1740
Query: 716 NSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGDHPTLYQALSEMGLLHR 775
NSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGDHPTLYQALSEMGLLHR
Sbjct: 1741 NSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGDHPTLYQALSEMGLLHR 1920
Query: 776 AVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIAAG 835
AVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIAAG
Sbjct: 1921 AVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIAAG 2100
Query: 836 KDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKTQGA 895
KDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKTQGA
Sbjct: 2101 KDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKTQGA 2280
Query: 896 AHVVVEIPSNMTESNKNPKQNESFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVY 955
AHVVVEIPSNMTESNKNPKQNESFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVY
Sbjct: 2281 AHVVVEIPSNMTESNKNPKQNESFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVY 2460
Query: 956 RPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGTS 999
RPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGTS
Sbjct: 2461 RPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGTS 2592
Score = 592 bits (1526), Expect(2) = 0.0
Identities = 306/342 (89%), Positives = 309/342 (89%), Gaps = 8/342 (2%)
Frame = +3
Query: 142 SNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFD 201
SNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFD
Sbjct: 3 SNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFD 182
Query: 202 EGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLISLLKILSNMHY----QPT 257
EGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLISLLKILSNMH QPT
Sbjct: 183 EGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLISLLKILSNMHSDRSDQPT 362
Query: 258 DQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPT 317
DQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPT
Sbjct: 363 DQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPT 542
Query: 318 VITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNF 377
VITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNF
Sbjct: 543 VITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNF 722
Query: 378 DLNDIYVDSDDGTEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSP- 436
DLNDIYVDSDDGTEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQ P
Sbjct: 723 DLNDIYVDSDDGTEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQVPF 902
Query: 437 ---SSSSGEAQSRTDRIVFKLFGKEPNEFPLVLRAQILDWLS 475
AQ+ + GK F L L +ILDWLS
Sbjct: 903 *F*RKKPRVAQT---ELFLNSSGKSQMNFLLYLEHRILDWLS 1019
>TC88329 similar to PIR|T47827|T47827 squamosa promoter binding protein-like
12 [imported] - Arabidopsis thaliana, partial (19%)
Length = 1277
Score = 533 bits (1373), Expect = e-151
Identities = 261/330 (79%), Positives = 296/330 (89%)
Frame = +3
Query: 670 LLESSETDPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQLKYRMVNLNSGVDLFPLQRFTW 729
LLESS+T PD EG GKI+AK+QAMDFIHEMGWLLHR Q+K V LNS +DLFPL RF W
Sbjct: 3 LLESSDTYPDNEGAGKIQAKNQAMDFIHEMGWLLHRRQIK-SSVRLNSSMDLFPLDRFKW 179
Query: 730 LMEFSMDHDWCAVVKKLLNLLLDETVNKGDHPTLYQALSEMGLLHRAVRRNSKQLVELLL 789
LMEFS+DHDWCAVVKKLLNL+LD TV+ GDH +LY ALSE+GLLHRAVRRNS+QLVELLL
Sbjct: 180 LMEFSVDHDWCAVVKKLLNLMLDGTVSTGDHTSLYLALSELGLLHRAVRRNSRQLVELLL 359
Query: 790 RYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTND 849
R+VP N S++LGPEDKALV G+N ++LFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTND
Sbjct: 360 RFVPQNISNKLGPEDKALVNGENQNFLFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTND 539
Query: 850 PCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKTQGAAHVVVEIPSNMTES 909
PCMVGIEAW +ARDSTGSTPEDYARLRGHYTYIHLVQKKINK+QG AHVVV+IPS T+
Sbjct: 540 PCMVGIEAWNSARDSTGSTPEDYARLRGHYTYIHLVQKKINKSQGGAHVVVDIPSIPTKF 719
Query: 910 NKNPKQNESFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVYRPAMLSMVAIAAVC 969
+ + K++ES T+ +IG AEV++ + +CKLCD K+SCRTAV +S VYRPAMLSMVAIAAVC
Sbjct: 720 DTSQKKDESCTTFQIGNAEVKKVRKDCKLCDHKLSCRTAVRKSFVYRPAMLSMVAIAAVC 899
Query: 970 VCVALLFKSSPEVLYMFRPFRWESLDFGTS 999
VCVALLFKSSPEVLY+FRPFRWESLD+GTS
Sbjct: 900 VCVALLFKSSPEVLYIFRPFRWESLDYGTS 989
>AW695484 similar to PIR|T52602|T526 squamosa promoter binding protein 1
[imported] - Arabidopsis thaliana, partial (8%)
Length = 468
Score = 228 bits (582), Expect = 7e-60
Identities = 111/154 (72%), Positives = 131/154 (84%), Gaps = 1/154 (0%)
Frame = +2
Query: 303 MVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAY 362
MVS L +NGSQGSPTV Q+Q VS++++Q +++H+HD R +D Q S KP +SNSPPAY
Sbjct: 5 MVSTLVTNGSQGSPTVTVQNQTVSISEIQHQVMHSHDARVADQQTTFSAKPGVSNSPPAY 184
Query: 363 SETRDSS-GQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLATSSVDYPWTQQDSHQSSP 421
SE R S+ GQTKMN+FDLNDIY+DSDDG ED+ERLPV+TNL SS+DYPW QQDSHQSSP
Sbjct: 185 SEARYSTAGQTKMNDFDLNDIYIDSDDGIEDIERLPVTTNLGASSLDYPWMQQDSHQSSP 364
Query: 422 AQTSGNSDSASAQSPSSSSGEAQSRTDRIVFKLF 455
QTSGNSDSASAQSPSSS+GE Q+RTDRIVFKLF
Sbjct: 365 PQTSGNSDSASAQSPSSSTGETQNRTDRIVFKLF 466
>TC78903
Length = 332
Score = 225 bits (573), Expect = 7e-59
Identities = 110/110 (100%), Positives = 110/110 (100%)
Frame = +3
Query: 25 GKRSREWNLNDWRWDGDLFIASRVNQVQAESLRVGQQFFPLGSGIPVVGGSSNTSSSCSE 84
GKRSREWNLNDWRWDGDLFIASRVNQVQAESLRVGQQFFPLGSGIPVVGGSSNTSSSCSE
Sbjct: 3 GKRSREWNLNDWRWDGDLFIASRVNQVQAESLRVGQQFFPLGSGIPVVGGSSNTSSSCSE 182
Query: 85 EGDLEKGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVSPVVERDGKK 134
EGDLEKGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVSPVVERDGKK
Sbjct: 183 EGDLEKGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVSPVVERDGKK 332
>TC80108 similar to GP|22045251|gb|AAL69483.2 putative SPL1-related protein
{Arabidopsis thaliana}, partial (42%)
Length = 1251
Score = 138 bits (347), Expect = 1e-32
Identities = 79/235 (33%), Positives = 127/235 (53%), Gaps = 1/235 (0%)
Frame = +2
Query: 765 QALSEMGLLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGP 824
+ L + LL+RAV+R +V+LL+ Y + +D + Y+F P+ GP
Sbjct: 23 EMLKAIQLLNRAVKRKCTSMVDLLINYSITSKNDT------------SKKYVFPPNLEGP 166
Query: 825 AGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHL 884
G+TPLH+AA SE V+D+LTNDP +G++ W+ D G TP YA +R +++Y L
Sbjct: 167 GGITPLHLAASTTDSEGVIDSLTNDPQEIGLKCWETLADENGQTPHAYAMMRNNHSYNML 346
Query: 885 VQKKINKTQGAAHVVVEIPSNMTESNKNPKQNESFTSLEIGKAE-VRRSQGNCKLCDTKI 943
V +K + Q + E+ + ++P I + + V S C + + +
Sbjct: 347 VARKCSDRQRS-----EVSVRIDNEIEHPSLGIELMQKRINQVKRVGDSCSKCAIAEVRA 511
Query: 944 SCRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGT 998
R + RS ++ P + SM+A+AAVCVCV +LF+ +P V PFRWE+L++GT
Sbjct: 512 KRRFSGSRSWLHGPFIHSMLAVAAVCVCVCVLFRGTPYV-GSVSPFRWENLNYGT 673
>TC82440 similar to PIR|H96793|H96793 unknown protein F14G6.18 [imported] -
Arabidopsis thaliana, partial (12%)
Length = 1244
Score = 109 bits (273), Expect(2) = 4e-30
Identities = 56/91 (61%), Positives = 67/91 (73%), Gaps = 1/91 (1%)
Frame = +1
Query: 115 ALSLNLAGHVSPVVERDGKKSRGAGGTS-NRAVCQVEDCGADLSRGKDYHRRHKVCEMHS 173
+L+LNL S + R K+ R TS + +CQV++C DLS+ KDYHRRHKVCE HS
Sbjct: 265 SLNLNLG---STGLVRPNKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHS 435
Query: 174 KASRALVGNAMQRFCQQCSRFHILEEFDEGK 204
KAS+AL+GN MQRFCQQCSRFH L EFDEGK
Sbjct: 436 KASKALLGNQMQRFCQQCSRFHPLVEFDEGK 528
Score = 41.2 bits (95), Expect(2) = 4e-30
Identities = 17/20 (85%), Positives = 19/20 (95%)
Frame = +2
Query: 202 EGKRSCRRRLAGHNKRRRKT 221
+G RSCRRRLAGHN+RRRKT
Sbjct: 521 KGNRSCRRRLAGHNRRRRKT 580
>BG455868 similar to SP|Q38741|SBP1_ Squamosa-promoter binding protein 1.
[Garden snapdragon] {Antirrhinum majus}, partial (70%)
Length = 656
Score = 123 bits (308), Expect = 4e-28
Identities = 58/92 (63%), Positives = 68/92 (73%)
Frame = +1
Query: 133 KKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCS 192
KK AGG+ + CQV++C ADLS K YH+RHKVCE HSKA L+ QRFCQQCS
Sbjct: 226 KKGSKAGGSVTPS-CQVDNCNADLSAAKQYHKRHKVCENHSKAHSVLISELQQRFCQQCS 402
Query: 193 RFHILEEFDEGKRSCRRRLAGHNKRRRKTNQE 224
RFH + EFD+ KRSCRRRLAGHN+RRRK+ E
Sbjct: 403 RFHEVSEFDDLKRSCRRRLAGHNERRRKSASE 498
>TC92499 homologue to PIR|T52594|T52594 squamosa promoter binding protein 8
[imported] - Arabidopsis thaliana, partial (22%)
Length = 805
Score = 122 bits (307), Expect = 5e-28
Identities = 57/84 (67%), Positives = 63/84 (74%)
Frame = +1
Query: 138 AGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHIL 197
A G S R CQ E C ADLS+ K YHRRHKVCE HSKA+ + QRFCQQCSRFH+L
Sbjct: 1 ARGNSPR--CQAEGCNADLSQAKHYHRRHKVCEFHSKAATVVAAGLTQRFCQQCSRFHLL 174
Query: 198 EEFDEGKRSCRRRLAGHNKRRRKT 221
EFD GKRSCR+RLA HN+RRRKT
Sbjct: 175 SEFDNGKRSCRKRLADHNRRRRKT 246
>AW691786 similar to PIR|T52298|T52 squamosa promoter binding protein-homolog
4 [imported] - garden snapdragon (fragment), partial
(42%)
Length = 649
Score = 116 bits (290), Expect = 5e-26
Identities = 55/89 (61%), Positives = 62/89 (68%)
Frame = +3
Query: 133 KKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCS 192
K+ G +S CQVE C DL+ K Y+ RHKVC MHSK+ V QRFCQQCS
Sbjct: 174 KEEVGQFNSSQPPRCQVEGCKLDLTDAKAYYSRHKVCSMHSKSPTVTVSGLQQRFCQQCS 353
Query: 193 RFHILEEFDEGKRSCRRRLAGHNKRRRKT 221
RFH L EFD+GKRSCRRRLAGHN+RRRKT
Sbjct: 354 RFHQLAEFDQGKRSCRRRLAGHNERRRKT 440
>CA916954 similar to PIR|T52592|T525 squamosa-promoter binding protein 6
[imported] - Arabidopsis thaliana, partial (16%)
Length = 775
Score = 103 bits (256), Expect = 4e-22
Identities = 47/72 (65%), Positives = 53/72 (73%)
Frame = +3
Query: 142 SNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFD 201
S A CQV C DLS KDYH+RHKVCE+HSK +V QRFCQQCSRFH+L EFD
Sbjct: 558 SMTAYCQVYGCNKDLSSCKDYHKRHKVCEVHSKTPIVIVNGIEQRFCQQCSRFHLLSEFD 737
Query: 202 EGKRSCRRRLAG 213
+G RSCR+RLAG
Sbjct: 738 DG*RSCRKRLAG 773
>BG582086 similar to PIR|T52603|T526 squamosa promoter binding protein 2
[imported] - Arabidopsis thaliana, partial (18%)
Length = 805
Score = 102 bits (254), Expect = 7e-22
Identities = 45/80 (56%), Positives = 59/80 (73%)
Frame = +1
Query: 147 CQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFDEGKRS 206
CQVE CG +LS KDYH + +VCE H+K+ ++ +RFCQQCSRFH L EFD K+S
Sbjct: 214 CQVEGCGLNLSSAKDYHCKRRVCESHAKSPMVVIDGLERRFCQQCSRFHDLFEFDGKKKS 393
Query: 207 CRRRLAGHNKRRRKTNQEAV 226
CRR+L+ HN RRRK +++AV
Sbjct: 394 CRRQLSNHNARRRKYHRQAV 453
>TC81138 similar to PIR|T52606|T52606 squamosa promoter binding protein 7
[imported] - Arabidopsis thaliana (fragment), partial
(32%)
Length = 743
Score = 97.4 bits (241), Expect = 2e-20
Identities = 43/89 (48%), Positives = 58/89 (64%)
Frame = +2
Query: 132 GKKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQC 191
GKK + A CQV C D+S K YHRRH+VC + A+ ++ ++R+CQQC
Sbjct: 383 GKKRARTARAAASARCQVPGCEVDISELKGYHRRHRVCLRCANAATVVLDGDVKRYCQQC 562
Query: 192 SRFHILEEFDEGKRSCRRRLAGHNKRRRK 220
+FH+L +FDEGKRSCRR+L HN RRR+
Sbjct: 563 GKFHVLSDFDEGKRSCRRKLERHNTRRRR 649
>TC91858 similar to PIR|T52592|T52592 squamosa-promoter binding protein 6
[imported] - Arabidopsis thaliana, partial (19%)
Length = 1130
Score = 91.7 bits (226), Expect = 1e-18
Identities = 45/89 (50%), Positives = 56/89 (62%)
Frame = +3
Query: 114 GALSLNLAGHVSPVVERDGKKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHS 173
G L+ +G +P K+ R + S A CQV C DLS KDYH+RHKVCE+HS
Sbjct: 879 GTTILSSSGSSTP-----SKRVRSSAIHSQIAYCQVYGCNKDLSSCKDYHKRHKVCEVHS 1043
Query: 174 KASRALVGNAMQRFCQQCSRFHILEEFDE 202
K S+ +V QRFCQQCSRFH+L EFD+
Sbjct: 1044KTSKVIVNGIEQRFCQQCSRFHLLAEFDD 1130
>TC78902
Length = 686
Score = 89.0 bits (219), Expect = 8e-18
Identities = 38/41 (92%), Positives = 38/41 (92%)
Frame = +1
Query: 1 MGERLGAENYHFYGVGGSSDLSGMGKRSREWNLNDWRWDGD 41
MGERLGAENYHFYGVGGSSDLSGMG SREWNLNDW WDGD
Sbjct: 562 MGERLGAENYHFYGVGGSSDLSGMGXXSREWNLNDWXWDGD 684
>BG587629 similar to PIR|T52605|T526 squamosa promoter binding protein 7
[imported] - Arabidopsis thaliana, partial (9%)
Length = 631
Score = 65.5 bits (158), Expect = 1e-10
Identities = 47/183 (25%), Positives = 80/183 (43%), Gaps = 18/183 (9%)
Frame = +2
Query: 566 VSPIAVPASKRAQFSVKGVNLMRPATRLMCALEGKYLVCEDAHESTDQYSEELDELQCI- 624
+ P A K +F G NL++P RL+ + GKYL CE S +E D + C
Sbjct: 101 IHPTCFEAGKPMEFFACGSNLLQPKFRLLVSFYGKYLKCEYCAPSPHNSAE--DNISCAF 274
Query: 625 ---QFSCSVPVSN----GRGFIEIEDQGLSSSFFPFIVAEEDVCTEIRVLEPLLESSETD 677
+ VP G FIE+E++ S+F P ++ ++++CTE+++L+ L++S
Sbjct: 275 DNQLYKICVPHIEENLLGPAFIEVENESGLSNFIPVLIGDKEICTELKILQQKLDASLLS 454
Query: 678 PDIEGTGKIKAKSQAMDFIH----------EMGWLLHRSQLKYRMVNLNSGVDLFPLQRF 727
S F+H ++ WLL + N + V +QR+
Sbjct: 455 KQFRSASGSSICSSCEAFVHIHTSSSDLLVDIAWLLKDPTSE----NFDRMVSASQIQRY 622
Query: 728 TWL 730
+L
Sbjct: 623 CYL 631
>TC92185 similar to GP|13374857|emb|CAC34491. putative protein {Arabidopsis
thaliana}, partial (5%)
Length = 557
Score = 56.2 bits (134), Expect = 6e-08
Identities = 31/67 (46%), Positives = 41/67 (60%), Gaps = 2/67 (2%)
Frame = +3
Query: 4 RLGAENYHFYGVGG-SSDLSGMGKRSREWNLNDWRWDGDLFIA-SRVNQVQAESLRVGQQ 61
R+ E + YG GG SSDL MGK S+EW+LN+W+WD LFIA S++ V +Q
Sbjct: 369 RVIMEAFQLYGNGGGSSDLRAMGKNSKEWDLNNWKWDSHLFIATSKLTPVPEH-----RQ 533
Query: 62 FFPLGSG 68
F P+ G
Sbjct: 534 FLPIPVG 554
>TC92276 similar to GP|15529228|gb|AAK97708.1 At1g03950/F21M11_12
{Arabidopsis thaliana}, partial (91%)
Length = 880
Score = 33.9 bits (76), Expect = 0.32
Identities = 23/70 (32%), Positives = 33/70 (46%), Gaps = 2/70 (2%)
Frame = +3
Query: 689 KSQAMDFIHEMGWLLHRSQLKYRMVNLNSGVDLFPLQRFTWLMEFSMDHDWCAV--VKKL 746
K+ F+ ++ W+LH S L V L G+ PLQ M ++ DW K L
Sbjct: 522 KNSLTRFLMKLEWILHPSYLLLPKVGLLQGMLKMPLQDQNPKMLKNLKRDWLLFEEYKDL 701
Query: 747 LNLLLDETVN 756
L LL+D +N
Sbjct: 702 LQLLIDMFLN 731
>TC84133 weakly similar to GP|22093574|dbj|BAC06871. contains ESTs
AU030377(E50973) AU030594(E51282)
AU030595(E51282)~similar to Arabidopsis thaliana,
partial (20%)
Length = 595
Score = 32.7 bits (73), Expect = 0.71
Identities = 25/92 (27%), Positives = 43/92 (46%), Gaps = 5/92 (5%)
Frame = +2
Query: 364 ETRDSSGQTKMNNFDLN-----DIYVDSDDGTEDLERLPVSTNLATSSVDYPWTQQDSHQ 418
E RD+ + FD+N D D DD +++++ P + + ++ ++ +
Sbjct: 131 EERDNVEEKDNVGFDMNVPLTDDEIADVDDSGDEVDKGPNAAEALRAHMNAEGREEQTST 310
Query: 419 SSPAQTSGNSDSASAQSPSSSSGEAQSRTDRI 450
SS + SG+S SAS SSSS S D +
Sbjct: 311 SSSSSGSGSS-SASGSGSSSSSDSDGSDEDSV 403
>TC88424 similar to GP|22136724|gb|AAM91681.1 unknown protein {Arabidopsis
thaliana}, partial (50%)
Length = 1635
Score = 32.3 bits (72), Expect = 0.93
Identities = 28/139 (20%), Positives = 59/139 (42%), Gaps = 8/139 (5%)
Frame = +2
Query: 196 ILEEFDEGKRSCRRR------LAGHNKRRRKTNQEAVPN--GSPTNDDQTSSYLLISLLK 247
+ E+ + KRS RR+ ++ + K++Q + P GS N+D +L++
Sbjct: 797 VQEDMTKNKRSLRRKSISPIDVSREEEGVSKSDQHSKPETPGSCVNNDSEHHRILVNFNA 976
Query: 248 ILSNMHYQPTDQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSAL 307
+ N+H + LL H+ S + + K N + + ++ + R S + +
Sbjct: 977 LTGNIHRDKWLERLLQHIQYMCNSDDSTEKEKGSGN---AESDEIKNKSNDRTSEIANDS 1147
Query: 308 FSNGSQGSPTVITQHQPVS 326
+ G + S + + VS
Sbjct: 1148QNKGQKSSESFVWPDAAVS 1204
>TC81895 weakly similar to PIR|T48577|T48577 hypothetical protein T31B5.120
- Arabidopsis thaliana, partial (16%)
Length = 853
Score = 32.0 bits (71), Expect = 1.2
Identities = 29/87 (33%), Positives = 44/87 (50%), Gaps = 9/87 (10%)
Frame = +2
Query: 772 LLHRAVRRNSKQLVELLLRYVPD-NTSDELG--PEDKALVGGKN---HSYLFR---PDAV 822
LLH A +VELLL+Y + N +D G P + ++ G++ L R P AV
Sbjct: 287 LLHIACETADIGMVELLLQYGANINATDMRGRTPLHRCILKGRSIIARLLLSRGGDPRAV 466
Query: 823 GPAGLTPLHIAAGKDGSEDVLDALTND 849
G TP+ +AA + + V+ A +ND
Sbjct: 467 DEDGRTPIELAAESNADDRVVHAPSND 547
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,876,207
Number of Sequences: 36976
Number of extensions: 441248
Number of successful extensions: 2500
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 2436
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2484
length of query: 999
length of database: 9,014,727
effective HSP length: 106
effective length of query: 893
effective length of database: 5,095,271
effective search space: 4550077003
effective search space used: 4550077003
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144728.1


