

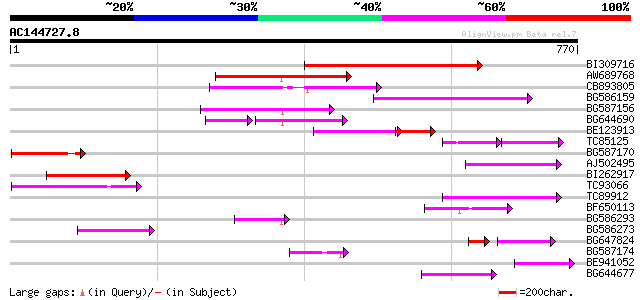
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144727.8 + phase: 0 /pseudo
(770 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 209 4e-54
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 182 3e-46
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 160 2e-39
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 154 1e-37
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 137 1e-32
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 97 4e-26
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 79 4e-23
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 62 4e-21
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 99 6e-21
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 98 1e-20
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 97 2e-20
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 97 2e-20
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 94 2e-19
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 85 9e-17
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 71 1e-12
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 67 3e-11
BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F... 52 1e-10
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 59 5e-09
BE941052 weakly similar to PIR|B85188|B85 retrotransposon like p... 58 1e-08
BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T... 57 3e-08
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 209 bits (531), Expect = 4e-54
Identities = 108/241 (44%), Positives = 154/241 (63%)
Frame = +2
Query: 401 VCKLKRSLYGLKQAPRAWYEKFCSSLLGFCFSHSQYNSSLFIHRTSTGIVLLLLYVDDMV 460
VC+L++S+YGLKQA R WY K SL+ F + S + SLF + LL+YVDD+V
Sbjct: 20 VCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDIV 199
Query: 461 ITGSDNAFIQRLKEQLHASFHMKDLGNLHYFLGLEVHSTSKGIFLHQHKYATYLISMAGL 520
+ G+D + IQ +K L F +KDLG+L YFLGLEV + +GI L+Q KY L+ +G
Sbjct: 200 LAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSGN 379
Query: 521 QSTNPVDTPLEVNVKYHRDDGDILPDPLLYRQLVGSLN*LTITRPDISFVVQQVSQFMHS 580
+ TP ++++K H D + D YR+L+G L LT TRPDISF VQQ+SQF+
Sbjct: 380 LAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVSK 559
Query: 581 PRHLHLAAVRRIIRYLKGSSHCGLFFSIGNYPKLSAYSDANWAGCPDTRHSVTSWCMFLR 640
P+ +H A R+++YLK + GLF+S + KLS+++D++WA CP TR SVT + +FL
Sbjct: 560 PQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFLG 739
Query: 641 S 641
S
Sbjct: 740 S 742
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 182 bits (463), Expect = 3e-46
Identities = 93/200 (46%), Positives = 127/200 (63%), Gaps = 15/200 (7%)
Frame = +1
Query: 280 CYSQVVKDVRWIKAMNEEIQALQENLTWDIVSCPPDIKPMGCKWVYSVKLNSDGSLNRYN 339
C + + D RW++AM E +AL +N TWD+V PP K +GCKWVY VK N DGS+N++
Sbjct: 67 CENLPLSDPRWLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFK 246
Query: 340 ARLVTLRNKQEYGIDYDETFAPVAKMTT--------------VHQMDVKNIFLHGDLAED 385
ARLV Q G DY ETF+PV K T + Q+D+ N FL+G L E+
Sbjct: 247 ARLVAKGFSQTLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEE 426
Query: 386 IYMTPPQGLFSSSKG-VCKLKRSLYGLKQAPRAWYEKFCSSLLGFCFSHSQYNSSLFIHR 444
+YM+ PQG +++K VCKL +SLYGLKQAPRAWYE S+ + F F+ S+ + SL I+
Sbjct: 427 VYMSQPQGFEAANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYN 606
Query: 445 TSTGIVLLLLYVDDMVITGS 464
+ + L +YVDD++ITGS
Sbjct: 607 QNGACIYLXIYVDDILITGS 666
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 160 bits (405), Expect = 2e-39
Identities = 88/268 (32%), Positives = 132/268 (48%), Gaps = 34/268 (12%)
Frame = +3
Query: 272 LSSISIPTCYSQVVKDVRWIKAMNEEIQALQENLTWDIVSCPPDIKPMGCKWVYSVKLNS 331
L+ S PT + + VK +W +MN E++A + N TW++ K +G KW++ KLN
Sbjct: 24 LTMTSDPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNE 203
Query: 332 DGSLNRYNARLVTLRNKQEYGIDYDETFAPVAKMTTVHQMDVKNIFLHGDLAEDIYMTPP 391
+G + +Y ARLV Q+YG+DY E FAPVA+ T+ + I L + D
Sbjct: 204 NGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMV----IALAAQIKRD------ 353
Query: 392 QGLFSSSKGVC---------------------------------KLKRSLYGLKQAPRAW 418
GVC ++KR+LYGLKQAPRAW
Sbjct: 354 --------GVCIS*M*KAHSCMEN*MRKFLLINHRVM*RRVIS*RVKRALYGLKQAPRAW 509
Query: 419 YEKFCSSLLGFCFSHSQYNSSLFIHRTSTGIVLLL-LYVDDMVITGSDNAFIQRLKEQLH 477
Y + + F Y +LF+ + G +L++ LYVDD++ G+D + K+ +
Sbjct: 510 YSRIEAYFTKEGFEKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMK 689
Query: 478 ASFHMKDLGNLHYFLGLEVHSTSKGIFL 505
F+M DLG +HYFLG+EV KGI++
Sbjct: 690 KEFNMSDLGKMHYFLGVEVTQNEKGIYI 773
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 154 bits (389), Expect = 1e-37
Identities = 77/217 (35%), Positives = 125/217 (57%)
Frame = +1
Query: 494 LEVHSTSKGIFLHQHKYATYLISMAGLQSTNPVDTPLEVNVKYHRDDGDILPDPLLYRQL 553
+EV +GI++ Q KY T L+ G++ +N P+ K +D+ + D Y+Q+
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 554 VGSLN*LTITRPDISFVVQQVSQFMHSPRHLHLAAVRRIIRYLKGSSHCGLFFSIGNYPK 613
VG L L TRPD+ +V+ +S+FM+ P LH+ AV+R++RYL G+ + G+ + K
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 614 LSAYSDANWAGCPDTRHSVTSWCMFLRSSLISWKSKKQARVSKSSTESEYRAMYAACSEI 673
L AY+D+++AG D R S + + L S +SW SKKQ V+ S+T++E+ A +
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 674 IWLRGFLAELGFP*TEPTSLYADNTSAI*FVANPVFH 710
+W+R L +LG+ + ++Y DN S I NPV H
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 137 bits (346), Expect = 1e-32
Identities = 74/198 (37%), Positives = 110/198 (55%), Gaps = 16/198 (8%)
Frame = -1
Query: 260 RYGSKHTSLTASLSSISIPTCYSQVVKDVRWIKAMNEEIQALQENLTWDIVSCPPDIKPM 319
+Y H + +L+ IP Y + ++D W +++ E A+ +N TW P K +
Sbjct: 612 QYPEAHCAFMVNLNENHIPRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAV 433
Query: 320 GCKWVYSVKLNSDGSLNRYNARLVTLRNKQEYGIDYDETFAPVAKMTTVH---------- 369
+W++++K +DGS+ R RLV YG DY ETFAPVAK+ T+
Sbjct: 432 SSRWIFTIKYKADGSIERKKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLG 253
Query: 370 ----QMDVKNIFLHGDLAEDIYMTPPQGLFSSSK--GVCKLKRSLYGLKQAPRAWYEKFC 423
QMDVKN FL G+L +++YM PP GL K V +LK+++YGLKQ+PRAWY K
Sbjct: 252 WGLWQMDVKNAFLQGELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLS 73
Query: 424 SSLLGFCFSHSQYNSSLF 441
++L G F S+ + +LF
Sbjct: 72 TTLNGRGFRKSELDHTLF 19
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 96.7 bits (239), Expect(2) = 4e-26
Identities = 51/141 (36%), Positives = 83/141 (58%), Gaps = 16/141 (11%)
Frame = -2
Query: 335 LNRYNARLVTLRNKQEYGIDYDETFAPVAKMTTV--------------HQMDVKNIFLHG 380
+ R ++LV Q+ GIDYDE F+PVA+M + +QMDVK+ F++G
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 381 DLAEDIYMTPPQGLFSSS--KGVCKLKRSLYGLKQAPRAWYEKFCSSLLGFCFSHSQYNS 438
DL E++++ P G + V +L ++LYGLKQAPRAWYE+ LL F + ++
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 439 SLFIHRTSTGIVLLLLYVDDM 459
+LF+ + ++++ +YVDD+
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 40.4 bits (93), Expect(2) = 4e-26
Identities = 20/63 (31%), Positives = 33/63 (51%)
Frame = -3
Query: 267 SLTASLSSISIPTCYSQVVKDVRWIKAMNEEIQALQENLTWDIVSCPPDIKPMGCKWVYS 326
S +A +SSI P + ++D WI +M EE+ + + W +V P +G +WV+
Sbjct: 615 SFSAFISSIE-PKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVFR 439
Query: 327 VKL 329
KL
Sbjct: 438 NKL 430
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 79.0 bits (193), Expect(2) = 4e-23
Identities = 44/122 (36%), Positives = 66/122 (54%), Gaps = 1/122 (0%)
Frame = +1
Query: 413 QAPRAWYEKFCSSLLGFCFSHSQYNSSLFIHRTST-GIVLLLLYVDDMVITGSDNAFIQR 471
Q+PR W+++F + F + Q + ++FI +ST +L++YVDD+ +TG I+R
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 472 LKEQLHASFHMKDLGNLHYFLGLEVHSTSKGIFLHQHKYATYLISMAGLQSTNPVDTPLE 531
LK L F +KDLGNL YFLG+EV KG + Q KY L+ + + P
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDPYG 360
Query: 532 VN 533
N
Sbjct: 361 CN 366
Score = 48.1 bits (113), Expect(2) = 4e-23
Identities = 27/54 (50%), Positives = 34/54 (62%)
Frame = +2
Query: 525 PVDTPLEVNVKYHRDDGDILPDPLLYRQLVGSLN*LTITRPDISFVVQQVSQFM 578
P +TP++ VK D L D Y++LVG L L+ TRPDISFVV +SQFM
Sbjct: 341 PSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 61.6 bits (148), Expect(2) = 4e-21
Identities = 29/83 (34%), Positives = 47/83 (55%)
Frame = +3
Query: 669 ACSEIIWLRGFLAELGFP*TEPTSLYADNTSAI*FVANPVFHERTKLIEVDCHSIRDAYD 728
AC E IW++ + ELG E ++Y D+ SA+ NP FH RTK I + H +R+ +
Sbjct: 240 ACKEAIWMQRLMEELGHK-QEQITVYCDSQSALHIARNPAFHSRTKHIGIQYHFVREVVE 416
Query: 729 E*LISLPHVSTQLQIADILTKAV 751
E + + + T +AD +TK++
Sbjct: 417 EGSVDMQKIHTNDNLADAMTKSI 485
Score = 58.5 bits (140), Expect(2) = 4e-21
Identities = 32/79 (40%), Positives = 47/79 (58%)
Frame = +1
Query: 589 VRRIIRYLKGSSHCGLFFSIGNYPKLSAYSDANWAGCPDTRHSVTSWCMFLRSSLISWKS 648
V+RI+RY+KG+S + F G+ + Y D+++AG D R S T + L +SW S
Sbjct: 1 VKRIMRYIKGTSGVAVCFG-GSELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLS 177
Query: 649 KKQARVSKSSTESEYRAMY 667
K Q V+ S+TE+EY A Y
Sbjct: 178 KLQTVVALSTTEAEYMAAY 234
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 99.0 bits (245), Expect = 6e-21
Identities = 48/101 (47%), Positives = 67/101 (65%)
Frame = -3
Query: 3 FKKFLTYVETQFQSSVKIFRSDSSGEYMSHEFQEYLQHKGILSQRSCPNTPQQNGLAERK 62
FK F YV + + +KI RSD+ GEY S+ F+ +L H GIL Q SCP TPQQNG+A+RK
Sbjct: 290 FKNFQAYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHGILHQTSCPYTPQQNGVAKRK 111
Query: 63 NRHLLDVTRSLLF*AFIPPRFWVEALATTVFLINRLPSIVI 103
N+HL++V RSL+F A + +T +LIN +P+ V+
Sbjct: 110 NKHLMEVARSLMFQANV---------STACYLINWIPTKVL 15
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 97.8 bits (242), Expect = 1e-20
Identities = 51/130 (39%), Positives = 75/130 (57%)
Frame = +2
Query: 620 ANWAGCPDTRHSVTSWCMFLRSSLISWKSKKQARVSKSSTESEYRAMYAACSEIIWLRGF 679
++WAG +TR S + + L + ISW SKKQ V+ S+ E+EY A + ++ +WLR
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 680 LAELGFP*TEPTSLYADNTSAI*FVANPVFHERTKLIEVDCHSIRDAYDE*LISLPHVST 739
L + PT +Y DN SAI NPVFH R+K I++ H IR+ E + + + T
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 740 QLQIADILTK 749
+ +IADI TK
Sbjct: 362 EEKIADIFTK 391
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 97.4 bits (241), Expect = 2e-20
Identities = 50/114 (43%), Positives = 72/114 (62%)
Frame = +1
Query: 51 NTPQQNGLAERKNRHLLDVTRSLLF*AFIPPRFWVEALATTVFLINRLPSIVIEFDSPFF 110
+TPQQNG+AER NR LL+ TR++L A + FW EA+ T ++INR PS VI+ +P
Sbjct: 82 HTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPME 261
Query: 111 HLFKFQLDYSDLHTFGCVCFVHLPPFEKHKLGVQSVQCAFMGYSNSHKGFVCYD 164
+DYS LH FGC +V E+ KL +S +C F+GY+++ KG+ +D
Sbjct: 262 MWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGYXLWD 423
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 97.1 bits (240), Expect = 2e-20
Identities = 63/180 (35%), Positives = 95/180 (52%), Gaps = 4/180 (2%)
Frame = +1
Query: 3 FKKFLTYVETQFQSSVKIFRSDSSGEYMSHEFQEYLQHKGILSQRSCPNTPQQNGLAERK 62
FKK+ VETQ +VK +D+ E+ S +F E+ + GI ++ P PQQNG+AER
Sbjct: 241 FKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERM 420
Query: 63 NRHLLDVTRSLLF*AFI--PPRFWVEALATTVFLINRLPSIVIEFDSPFFHLFKFQLDYS 120
R LL+ R +L A + WVEA +T L+NR P ++F P +DYS
Sbjct: 421 IRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYS 600
Query: 121 DLHTFGCVCFVHLPPFEKHKLGVQSVQCAFMGYSNSHKGF--VCYDVSNHSFRVSRNVTF 178
+L FGC + + KL ++ +C F+ Y++ KG+ C D + +SR+VTF
Sbjct: 601 NLRIFGCPAYALV---NDGKLAPRAGECIFLSYASESKGYRLWCSDPKSQKLILSRDVTF 771
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 94.4 bits (233), Expect = 2e-19
Identities = 55/164 (33%), Positives = 93/164 (56%), Gaps = 2/164 (1%)
Frame = +1
Query: 588 AVRRIIRYLKGSSHCGLFFSIGNYPK--LSAYSDANWAGCPDTRHSVTSWCMFLRSSLIS 645
A++ +++YL S L ++ + L Y DA++AG DTR S++ + L + IS
Sbjct: 4 ALKWVLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTIS 183
Query: 646 WKSKKQARVSKSSTESEYRAMYAACSEIIWLRGFLAELGFP*TEPTSLYADNTSAI*FVA 705
WK+ +Q+ V+ S+T++EY A + IWL+G + ELG E ++ D+ SAI
Sbjct: 184 WKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGIT-QEYVKIHCDSQSAIHLAN 360
Query: 706 NPVFHERTKLIEVDCHSIRDAYDE*LISLPHVSTQLQIADILTK 749
+ V+HERTK I++ H IRD + I + ++++ AD+ TK
Sbjct: 361 HQVYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTK 492
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 85.1 bits (209), Expect = 9e-17
Identities = 48/123 (39%), Positives = 67/123 (54%), Gaps = 3/123 (2%)
Frame = +1
Query: 564 RPDISFVVQQVSQFMHSPRHLHLAAVRRIIRYLKGSSHCGLFFSIG---NYPKLSAYSDA 620
RPDI + V +S+FMH PR HL A RI+RY++G+ GL F G +L YSD+
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 621 NWAGCPDTRHSVTSWCMFLRSSLISWKSKKQARVSKSSTESEYRAMYAACSEIIWLRGFL 680
+W G R S + + + ISW +KKQ + SS E+EY A A + +WL +
Sbjct: 301 DWCG---DRRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 681 AEL 683
EL
Sbjct: 472 KEL 480
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 71.2 bits (173), Expect = 1e-12
Identities = 35/89 (39%), Positives = 51/89 (56%), Gaps = 14/89 (15%)
Frame = +2
Query: 306 TWDIVSCPPDIKPMGCKWVYSVKLNSDGSLNRYNARLVTLRNKQEYGIDYDETFAPVAKM 365
T +V P +KP+G +W+Y +K N DG+L +Y ARLV ++ GID+DE FAPV ++
Sbjct: 56 TLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPVVRI 235
Query: 366 TT--------------VHQMDVKNIFLHG 380
T +H +DVK FL+G
Sbjct: 236 ETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 66.6 bits (161), Expect = 3e-11
Identities = 35/105 (33%), Positives = 60/105 (56%), Gaps = 1/105 (0%)
Frame = -2
Query: 93 FLINRLPSIVIEFDSPFFHLFKFQLDYSDLHTFGCVCFVHLPPFEKHKLGVQSVQCAFMG 152
+LINR+P+ V++ +PF L + + + + FGC+C+V +P ++KL +S + F+G
Sbjct: 698 YLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMFIG 519
Query: 153 YSNSHKGFVCYDVSNHSFRVSRNVTFFYNQ-FMLHSISPDINDIT 196
YS + KG+ CYD VSR+V F + + D+ D+T
Sbjct: 518 YSTTQKGYKCYDPEARRVLVSRDVKFIEERGYYEEKNQEDLRDLT 384
>BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (5%)
Length = 721
Score = 52.4 bits (124), Expect(2) = 1e-10
Identities = 32/80 (40%), Positives = 43/80 (53%), Gaps = 1/80 (1%)
Frame = -3
Query: 663 YRAMYAACSEIIWLRGFLAELGFP*TEPTSLYADNTSAI*FV-ANPVFHERTKLIEVDCH 721
YR++ + EI WL L +L F +P LY DN SA + AN F ERTK IE+DCH
Sbjct: 374 YRSI*STICEIKWLTYLLNDLKFTFIKPAMLYCDNQSAARHIAANSSFLERTKHIELDCH 195
Query: 722 SIRDAYDE*LISLPHVSTQL 741
+R L + H+ + L
Sbjct: 194 IVRVKLQLKLFHILHILSSL 135
Score = 32.0 bits (71), Expect(2) = 1e-10
Identities = 14/29 (48%), Positives = 20/29 (68%)
Frame = -2
Query: 623 AGCPDTRHSVTSWCMFLRSSLISWKSKKQ 651
+ C DT S++ +C+FL SLI WKS K+
Sbjct: 480 SSCLDT*KSISYFCIFLGDSLICWKS*KK 394
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse
transcriptase homolog - rape retrotransposon copia-like
(fragment), partial (84%)
Length = 249
Score = 59.3 bits (142), Expect = 5e-09
Identities = 36/89 (40%), Positives = 50/89 (55%), Gaps = 9/89 (10%)
Frame = -1
Query: 381 DLAEDIYMTPPQGLFSSSKG--VCKLKRSLYGLKQAPRAWYEKFCSSLLGFCFSHSQYNS 438
+L E IYMT P+G K VCKL++SLYGLKQ+PR WY++F S + S +
Sbjct: 249 ELEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRFDS------YRSSWATT 88
Query: 439 SLFIHRTST-------GIVLLLLYVDDMV 460
+ + ST + L+LYVDDM+
Sbjct: 87 GVLMTVVST*TR*RMSRYIYLVLYVDDML 1
>BE941052 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 480
Score = 58.2 bits (139), Expect = 1e-08
Identities = 31/82 (37%), Positives = 44/82 (52%)
Frame = +2
Query: 686 P*TEPTSLYADNTSAI*FVANPVFHERTKLIEVDCHSIRDAYDE*LISLPHVSTQLQIAD 745
P ++ L D SA NPV+H R K I +D H +RD + + + HV T Q+AD
Sbjct: 2 PISQSGLLRCDYLSATYLTHNPVYHSRMKHISIDIHFVRDLVQQGKLKVQHVCTVDQLAD 181
Query: 746 ILTKAVPRPRHQFLVGKLMIFD 767
LTK + + RHQ L K+ + D
Sbjct: 182 CLTKPLSKSRHQLLRNKIGVTD 247
>BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T15F17.l
{Arabidopsis thaliana}, partial (3%)
Length = 539
Score = 56.6 bits (135), Expect = 3e-08
Identities = 31/102 (30%), Positives = 54/102 (52%)
Frame = -3
Query: 560 LTITRPDISFVVQQVSQFMHSPRHLHLAAVRRIIRYLKGSSHCGLFFSIGNYPKLSAYSD 619
LT+ P+I+F + +S++ +P H ++ I +YLKG GLF+S P L Y +
Sbjct: 525 LTLQGPNITFSINLLSRYSSAPTMRH*NGIKHICKYLKGIIDMGLFYSKDCSPDLIGYVN 346
Query: 620 ANWAGCPDTRHSVTSWCMFLRSSLISWKSKKQARVSKSSTES 661
A + P S T + +++ISW+S K + ++ SS +
Sbjct: 345 A*YLSDPHKARS*TGYIFTCGNTVISWRSTK*STIATSSNHA 220
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.139 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,674,128
Number of Sequences: 36976
Number of extensions: 484448
Number of successful extensions: 5685
Number of sequences better than 10.0: 88
Number of HSP's better than 10.0 without gapping: 3231
Number of HSP's successfully gapped in prelim test: 150
Number of HSP's that attempted gapping in prelim test: 863
Number of HSP's gapped (non-prelim): 4548
length of query: 770
length of database: 9,014,727
effective HSP length: 104
effective length of query: 666
effective length of database: 5,169,223
effective search space: 3442702518
effective search space used: 3442702518
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144727.8


