

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144727.5 + phase: 0
(256 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
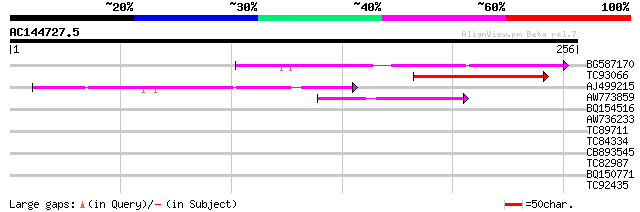
Score E
Sequences producing significant alignments: (bits) Value
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 80 1e-15
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 64 4e-11
AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 50 1e-06
AW773859 48 4e-06
BQ154516 36 0.017
AW736233 GP|21105451|gb small nuclear ribonucleoprotein D1 {Dani... 30 0.70
TC89711 weakly similar to PIR|T10725|T10725 protein kinase Xa21 ... 30 1.2
TC84334 weakly similar to PIR|T49308|T49308 hypothetical protein... 29 2.0
CB893545 similar to GP|8778537|gb| F22G5.5 {Arabidopsis thaliana... 27 5.9
TC82987 similar to GP|15991740|gb|AAL13047.1 WRKY transcription ... 27 7.7
BQ150771 GP|4028979|emb glutamate permease {synthetic construct}... 27 7.7
TC92435 similar to GP|18377843|gb|AAL67108.1 AT5g42350/MDH9_4 {A... 27 7.7
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 79.7 bits (195), Expect = 1e-15
Identities = 49/154 (31%), Positives = 77/154 (49%), Gaps = 4/154 (2%)
Frame = -3
Query: 103 SGKVIAKGPKVGRLFPLQF---ISNH-LSLPCNNVLNSYEDWHRKLGHPNSTVLSHLFKT 158
S ++I +G G L+ L+ +SN+ S ++ LN WH +LGHP+ L+ +
Sbjct: 704 SSQLIGEGVTKGDLYMLEKLDPVSNYKCSFTSSSSLNKDALWHARLGHPHGRALNLMLPG 525
Query: 159 GLLGNKQVVCTASISCLVCKLAKSKTLPFPSGAHRASNCFEMIHSDVWGMSPIASHARYK 218
+ NK +C C L K FP + NCF++I++D+W +P S +K
Sbjct: 524 VVFENK--------NCEACILGKHCKNVFPRTSTVYENCFDLIYTDLW-TAPSLSRDNHK 372
Query: 219 YFVTFIDDYSRFTWIYFLRSKSEVFSIVITISGY 252
YFVTFID+ S++TW+ + SK V Y
Sbjct: 371 YFVTFIDEKSKYTWLTLIPSKDRVIDAFKNFQAY 270
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 64.3 bits (155), Expect = 4e-11
Identities = 26/61 (42%), Positives = 37/61 (60%)
Frame = +1
Query: 183 KTLPFPSGAHRASNCFEMIHSDVWGMSPIASHARYKYFVTFIDDYSRFTWIYFLRSKSEV 242
K + F + HR + IHSD+WG S + S+ +Y +T IDD+ R W+YFLR K+E
Sbjct: 52 KKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNET 231
Query: 243 F 243
F
Sbjct: 232 F 234
>AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (18%)
Length = 567
Score = 49.7 bits (117), Expect = 1e-06
Identities = 37/154 (24%), Positives = 76/154 (49%), Gaps = 7/154 (4%)
Frame = +3
Query: 11 SRPWFLDSGASNHMTGSSEYLHNLASYHGNQQIQIADGNNLSITDVGDI----NSDFR-- 64
S+ W +DSG ++HMT + L + ++++ +G ++ + +G + +S ++
Sbjct: 99 SKYWLIDSGCTHHMTHDRDLFKEL-NKSTISKVRMLNGAHIEVEGIGTVLVKSHSGYKQI 275
Query: 65 -NVLVSPGLASNLLSVGQLVDNNCNVNFSRAGCVVQEQVSGKVIAKGPKVGRLFPLQFIS 123
NVL +P L +LLSV QL+ V F CV+++Q + K + + R F L +
Sbjct: 276 SNVLYAPKLNQSLLSVPQLLTKGYKVLFEHEKCVIKDQ-NNKEVLRAQMKARKFVLDLKN 452
Query: 124 NHLSLPCNNVLNSYEDWHRKLGHPNSTVLSHLFK 157
+++ + ++++ K H ++ VL K
Sbjct: 453 GYIT----DEDEKKKEYNFKEDHNDACVLKKFIK 542
>AW773859
Length = 538
Score = 47.8 bits (112), Expect = 4e-06
Identities = 22/68 (32%), Positives = 34/68 (49%)
Frame = -3
Query: 140 WHRKLGHPNSTVLSHLFKTGLLGNKQVVCTASISCLVCKLAKSKTLPFPSGAHRASNCFE 199
WH +LGH ++ L L + + C +C ++ K LPF +RAS C+E
Sbjct: 227 WHFRLGHLSNRKLLSLHSNFPF----ITIDQNSVCDICHYSRHKKLPFQLSTNRASKCYE 60
Query: 200 MIHSDVWG 207
+ H D+WG
Sbjct: 59 LFHFDIWG 36
>BQ154516
Length = 506
Score = 35.8 bits (81), Expect = 0.017
Identities = 16/42 (38%), Positives = 25/42 (59%)
Frame = +3
Query: 7 SSNVSRPWFLDSGASNHMTGSSEYLHNLASYHGNQQIQIADG 48
SS PW+ DSGAS+H+T + L Y+G +Q+ + +G
Sbjct: 105 SSYSEAPWYPDSGASHHLTFNPHNLAYRTPYNGQEQVLMDNG 230
>AW736233 GP|21105451|gb small nuclear ribonucleoprotein D1 {Danio rerio},
partial (14%)
Length = 510
Score = 30.4 bits (67), Expect = 0.70
Identities = 14/33 (42%), Positives = 19/33 (57%)
Frame = +1
Query: 7 SSNVSRPWFLDSGASNHMTGSSEYLHNLASYHG 39
SS PW+ DSGAS+H+T + L Y+G
Sbjct: 412 SSYSEAPWYPDSGASHHLTFNPHNLAYRTPYNG 510
>TC89711 weakly similar to PIR|T10725|T10725 protein kinase Xa21 (EC
2.7.1.-) A1 receptor type - long-staminate rice,
partial (7%)
Length = 1391
Score = 29.6 bits (65), Expect = 1.2
Identities = 11/22 (50%), Positives = 13/22 (59%)
Frame = -2
Query: 186 PFPSGAHRASNCFEMIHSDVWG 207
PF S A E+IH+DVWG
Sbjct: 904 PFKSSTSHAQEVLELIHTDVWG 839
>TC84334 weakly similar to PIR|T49308|T49308 hypothetical protein T16L24.220
- Arabidopsis thaliana, partial (14%)
Length = 804
Score = 28.9 bits (63), Expect = 2.0
Identities = 17/60 (28%), Positives = 30/60 (49%)
Frame = +1
Query: 169 TASISCLVCKLAKSKTLPFPSGAHRASNCFEMIHSDVWGMSPIASHARYKYFVTFIDDYS 228
TA+ K + SK+LPF S +R+ V + IAS+A + Y +++++ S
Sbjct: 514 TAAHDHFTLKESGSKSLPFSSHQYRSKAKMRRRRKKVEDTTDIASYAAHHYLFSYLENRS 693
>CB893545 similar to GP|8778537|gb| F22G5.5 {Arabidopsis thaliana}, partial
(27%)
Length = 824
Score = 27.3 bits (59), Expect = 5.9
Identities = 14/48 (29%), Positives = 25/48 (51%)
Frame = -2
Query: 135 NSYEDWHRKLGHPNSTVLSHLFKTGLLGNKQVVCTASISCLVCKLAKS 182
N +WH+K + V H F+T L + +C+A ++C C + +S
Sbjct: 562 NDLFNWHQK----TNLVSPHAFETRLRRG*EEICSAEVTC--CNVLRS 437
>TC82987 similar to GP|15991740|gb|AAL13047.1 WRKY transcription factor 71
{Arabidopsis thaliana}, partial (33%)
Length = 975
Score = 26.9 bits (58), Expect = 7.7
Identities = 15/52 (28%), Positives = 23/52 (43%), Gaps = 11/52 (21%)
Frame = +1
Query: 21 SNHMTGSSEYLHNLASYH-----------GNQQIQIADGNNLSITDVGDINS 61
S+H T S+YLH Y+ ++ I D NN + GD++S
Sbjct: 301 SSHSTSFSDYLHGSMDYNTLSKAFDLSCSSSEVISSIDDNNTKKSSAGDLSS 456
>BQ150771 GP|4028979|emb glutamate permease {synthetic construct}, partial
(3%)
Length = 1234
Score = 26.9 bits (58), Expect = 7.7
Identities = 11/32 (34%), Positives = 18/32 (55%)
Frame = -3
Query: 157 KTGLLGNKQVVCTASISCLVCKLAKSKTLPFP 188
++GLL + +++C +SCL TLP P
Sbjct: 476 RSGLLASGRLLCLCFVSCLCIHFFSLHTLPAP 381
>TC92435 similar to GP|18377843|gb|AAL67108.1 AT5g42350/MDH9_4 {Arabidopsis
thaliana}, partial (19%)
Length = 611
Score = 26.9 bits (58), Expect = 7.7
Identities = 14/35 (40%), Positives = 23/35 (65%), Gaps = 1/35 (2%)
Frame = -1
Query: 122 ISNHLSLPCNNV-LNSYEDWHRKLGHPNSTVLSHL 155
+ +H L CN+V L +YED HR+L S++L ++
Sbjct: 269 LPSH*RLICNDVSLKAYEDPHRRLIPHQSSILENV 165
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.135 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,101,316
Number of Sequences: 36976
Number of extensions: 138310
Number of successful extensions: 741
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 732
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 738
length of query: 256
length of database: 9,014,727
effective HSP length: 94
effective length of query: 162
effective length of database: 5,538,983
effective search space: 897315246
effective search space used: 897315246
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC144727.5


