

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144727.12 - phase: 0
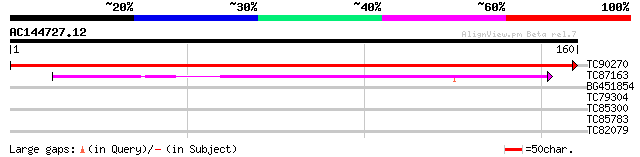
(160 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC90270 similar to PIR|B86418|B86418 hypothetical protein AAG126... 312 3e-86
TC87163 similar to PIR|T07661|T07661 maturation protein PM3 - so... 113 4e-26
BG451854 similar to SP|P82538|TL26 Thylakoid lumenal 25.6 kDa pr... 29 0.72
TC79304 similar to GP|9759186|dbj|BAB09801.1 emb|CAB66399.1~gene... 27 4.6
TC85300 homologue to PIR|S33775|S33775 chlorophyll a/b-binding p... 26 6.1
TC85783 homologue to GP|15131688|emb|CAC48323. 2-Cys peroxiredox... 26 7.9
TC82079 weakly similar to GP|11994399|dbj|BAB02358. contains sim... 26 7.9
>TC90270 similar to PIR|B86418|B86418 hypothetical protein AAG12626.1
[imported] - Arabidopsis thaliana, partial (97%)
Length = 789
Score = 312 bits (800), Expect = 3e-86
Identities = 158/160 (98%), Positives = 158/160 (98%)
Frame = +1
Query: 1 MANEQMKPIATLLLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMG 60
MANEQMKPIATLLLALNFCMYVIVISIGGWAMNRAIDHGFEI P FDLPAHFSPIYFPMG
Sbjct: 76 MANEQMKPIATLLLALNFCMYVIVISIGGWAMNRAIDHGFEISPRFDLPAHFSPIYFPMG 255
Query: 61 NAATGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCK 120
NAATGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCK
Sbjct: 256 NAATGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCK 435
Query: 121 EIQLNIRNSRLKTMEAFLIILTATQLFYIAAIHGAAAIRR 160
EIQLNIRNSRLKTMEAFLIILTATQLFYIAAIHGAAAIRR
Sbjct: 436 EIQLNIRNSRLKTMEAFLIILTATQLFYIAAIHGAAAIRR 555
>TC87163 similar to PIR|T07661|T07661 maturation protein PM3 - soybean,
partial (93%)
Length = 865
Score = 113 bits (282), Expect = 4e-26
Identities = 65/142 (45%), Positives = 83/142 (57%), Gaps = 1/142 (0%)
Frame = +1
Query: 13 LLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFAL 72
LL LN MY IV+ W +NR I+ G P F GN AT FF+TF++
Sbjct: 154 LLFLNLVMYFIVLGFASWCLNRFIN-GQTYHPSFG------------GNGATMFFLTFSI 294
Query: 73 IAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCKEIQL-NIRNSRL 131
+A V+GI S G NH+R W S+SL SA + + IAWA+T LAMG CK+I + R RL
Sbjct: 295 LAAVLGIVSKFMGANHLRFWRSDSLASAGATSVIAWAVTALAMGLACKQIHIGGHRGWRL 474
Query: 132 KTMEAFLIILTATQLFYIAAIH 153
K +EAF+IILT TQL + H
Sbjct: 475 KMVEAFIIILTFTQLLCLLLTH 540
>BG451854 similar to SP|P82538|TL26 Thylakoid lumenal 25.6 kDa protein
chloroplast precursor. [Mouse-ear cress] {Arabidopsis
thaliana}, partial (49%)
Length = 636
Score = 29.3 bits (64), Expect = 0.72
Identities = 16/60 (26%), Positives = 25/60 (41%)
Frame = -3
Query: 82 LISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCKEIQLNIRNSRLKTMEAFLIIL 141
L+S I W + A + AIAW L V C + + S L+ + L++L
Sbjct: 316 LLSAAKEIEDWLNRKTNGAVAAMAIAWRLPVEEDESSCSALTMKEMPSLLRKVLLLLLLL 137
>TC79304 similar to GP|9759186|dbj|BAB09801.1
emb|CAB66399.1~gene_id:K19E1.19~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (8%)
Length = 790
Score = 26.6 bits (57), Expect = 4.6
Identities = 14/53 (26%), Positives = 23/53 (42%)
Frame = -2
Query: 70 FALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCKEI 122
++LIA + S L I+ W S S+ + W+ T+ + GC I
Sbjct: 321 YSLIADSSSLSSETDSLLMIKMWRSRSILIGRQCSTPRWSRTMRLVNLGCWSI 163
>TC85300 homologue to PIR|S33775|S33775 chlorophyll a/b-binding protein -
garden pea, partial (96%)
Length = 1163
Score = 26.2 bits (56), Expect = 6.1
Identities = 13/29 (44%), Positives = 16/29 (54%)
Frame = -2
Query: 80 GSLISGLNHIRSWTSESLPSAASVAAIAW 108
GSL S L H S + PSA S+A + W
Sbjct: 538 GSLKSTLTHFSSTSGVMHPSAPSIAHLPW 452
>TC85783 homologue to GP|15131688|emb|CAC48323. 2-Cys peroxiredoxin {Pisum
sativum}, partial (91%)
Length = 1136
Score = 25.8 bits (55), Expect = 7.9
Identities = 18/59 (30%), Positives = 27/59 (45%), Gaps = 3/59 (5%)
Frame = -2
Query: 42 IGPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIGSL---ISGLNHIRSWTSESL 97
+G GF FSP + P G ++GF T+ V+ + S I L + W + SL
Sbjct: 814 LGSGF---MDFSPGFHPAGQTSSGFSCTYCKAWRVLFVSSTLLPIPRLLMVECWITPSL 647
>TC82079 weakly similar to GP|11994399|dbj|BAB02358. contains similarity to
receptor protein kinase~gene_id:MIL23.20 {Arabidopsis
thaliana}, partial (23%)
Length = 1057
Score = 25.8 bits (55), Expect = 7.9
Identities = 18/47 (38%), Positives = 23/47 (48%)
Frame = +2
Query: 33 NRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGI 79
N +I G PG D HF P+Y G A G V A+ AG+ G+
Sbjct: 623 NFSIGSGIVFIPGRDQNGHFFPLYSRTG-IAKGSAVGIAM-AGIFGL 757
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.328 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,773,286
Number of Sequences: 36976
Number of extensions: 61434
Number of successful extensions: 392
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 387
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 391
length of query: 160
length of database: 9,014,727
effective HSP length: 88
effective length of query: 72
effective length of database: 5,760,839
effective search space: 414780408
effective search space used: 414780408
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 54 (25.4 bits)
Medicago: description of AC144727.12


