

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144724.7 - phase: 0 /pseudo
(1419 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
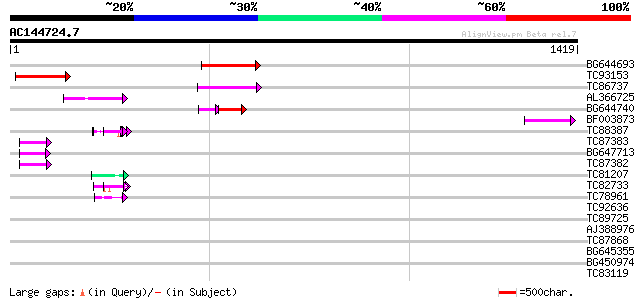
Score E
Sequences producing significant alignments: (bits) Value
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 152 7e-37
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 150 4e-36
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 132 1e-30
AL366725 117 3e-26
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 81 1e-20
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 79 2e-14
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 60 5e-09
TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA PO... 57 4e-08
BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent vir... 54 6e-07
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 52 2e-06
TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 51 3e-06
TC82733 similar to GP|10177404|dbj|BAB10535. gene_id:K24M7.12~pi... 50 6e-06
TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 47 5e-05
TC92636 homologue to GP|15042313|gb|AAK82093.1 232R {Chilo iride... 35 0.001
TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.... 37 0.042
AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing fact... 37 0.071
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 35 0.16
BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.... 35 0.21
BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR p... 34 0.46
TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 34 0.46
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 152 bits (385), Expect = 7e-37
Identities = 79/149 (53%), Positives = 102/149 (68%)
Frame = +2
Query: 480 LPLEREVEFAIDLVPGMSPIWITPYPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVL 539
+P E +++F IDL+P M+PI I Y ++ +L LK QL++LLEK FI+PS+ P G VL
Sbjct: 17 VPPEWKIDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVVVL 196
Query: 540 LVKKKDGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVK 599
+KKKDG +R+ +DY QLN V IK +YPL ID+L D L G++ F KID R G H +RV
Sbjct: 197 FLKKKDGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVI 376
Query: 600 AEDISKTVFRTRYGHYEYFVMQFGVYNVP 628
ED+ KT FR RYGHYE VM FG N P
Sbjct: 377 GEDVPKTAFRIRYGHYEILVMSFG*TNPP 463
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 150 bits (378), Expect = 4e-36
Identities = 74/138 (53%), Positives = 96/138 (68%)
Frame = +2
Query: 15 RRLDRFLRNNPPTYNGRFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTTHMLAEEAEYWW 74
R L+ FLRN+PPT+ GR+ P+GA W++ +ERIF M + QKV+ THMLAEEA+ WW
Sbjct: 101 RMLETFLRNHPPTFKGRYAPDGA*KWLKEIERIFRVMQCFETQKVQFGTHMLAEEADDWW 280
Query: 75 TNAKGRLEIVGEVVTWAKFKAEFLRKYFPEDLRTRKEVAFLNLK*GSISVAEYAAKFEKL 134
+ LE VVTWA F+ EFL +YFPED+R +KE+ FL LK G +SV EYAAKF +L
Sbjct: 281 ISLLPVLEQDDAVVTWAMFRKEFLGRYFPEDVRGKKEIEFLELKQGDMSVTEYAAKFVEL 460
Query: 135 SRFCPYIIAEDAMVSKCV 152
+ F P+ AE A SKC+
Sbjct: 461 ATFYPHYSAETAEFSKCI 514
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 132 bits (331), Expect = 1e-30
Identities = 77/173 (44%), Positives = 104/173 (59%), Gaps = 11/173 (6%)
Frame = +1
Query: 469 IVQEFP-----EDITELPLEREV-EFAIDLVP---GMSPI--WITPYPMSASELGELKKQ 517
+++EFP E ++P R + + AI L+P G P W Y MS EL LKK
Sbjct: 343 VLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKT 522
Query: 518 LEELLEKQFIRPSVSPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQ 577
LE+LL+K FI+ S S GAPVL V+K G +R CVDYR LN +T K+RYPL I + + +
Sbjct: 523 LEDLLDKGFIKASGSAAGAPVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLRR 702
Query: 578 LVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYNVPGT 630
+ GA F+K+D + +H R+K ED KT FRTRYG +E+ V FG+ P T
Sbjct: 703 VAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPAT 861
>AL366725
Length = 485
Score = 117 bits (293), Expect = 3e-26
Identities = 66/166 (39%), Positives = 99/166 (58%), Gaps = 7/166 (4%)
Frame = +2
Query: 136 RFCPYIIAEDAMVSKCVKFESGLRPDIYQYMCVQEIRDFDTLVHKCRKFDDAGRVKANYY 195
+F P+ AE A SKC+KFE+GLRPDI + + Q++R F LV+ CR +++ + +
Sbjct: 2 KFYPHYAAETAEFSKCIKFENGLRPDIKRAIGYQQLRVFPDLVNTCRIYEEDTKA---HD 172
Query: 196 KAQSEKRGKGH-GVGKPYN--KDKRKKREVGG---GSKPSLADIKCYRCGTLGHYANDCK 249
K +E++ KG KPY+ DK K+R V K + A+I C+ G GH +N C
Sbjct: 173 KVVNERKTKGQ*SRPKPYSAPADKGKQRMVDDRRPKKKDAPAEIVCFNYGEKGHKSNVCP 352
Query: 250 KDI-SCHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTKCTKPKK 294
K+I C +C K GH A+CK D+ C+NC E+GHI ++C +PK+
Sbjct: 353 KEIKKCVRCDKKGHIVADCK--RNDIVCFNCNEEGHIGSQCKQPKR 484
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 80.9 bits (198), Expect(2) = 1e-20
Identities = 39/69 (56%), Positives = 50/69 (71%)
Frame = -1
Query: 524 KQFIRPSVSPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEV 583
K+F +PS+SP GA +L V+KKDG R+C+DYRQ NKVT KN+YPL RID+L D++
Sbjct: 274 KRFQQPSISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCY 95
Query: 584 FSKIDFRSG 592
F ID R G
Sbjct: 94 F*NIDLRLG 68
Score = 38.9 bits (89), Expect(2) = 1e-20
Identities = 23/54 (42%), Positives = 32/54 (58%)
Frame = -2
Query: 473 FPEDITELPLEREVEFAIDLVPGMSPIWITPYPMSASELGELKKQLEELLEKQF 526
FP++ +PLERE+ F IDL+ I P M +EL +LK L++ LEK F
Sbjct: 426 FPDNFPVIPLEREIFFCIDLLLDTQLISNPP*LMDRTELKKLKI*LKDSLEKGF 265
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 78.6 bits (192), Expect = 2e-14
Identities = 52/129 (40%), Positives = 66/129 (50%)
Frame = +1
Query: 1288 WSWSYFEV*EVDSSFCWAV*CC*DGWGCSVSDCVTIVFVESS*CVSCVSVEEICVRCVSC 1347
WSW+ FEV EVD W+V W VS +T E +*C+SCV+ E+C +SC
Sbjct: 1 WSWTCFEVKEVDCEIHWSVSDIRKSWNGGVSSGITTASFEFA*CLSCVATSEVCSGSISC 180
Query: 1348 DSSGRIGSKR*FDR*DLAG*D*GP*SEAFAWQGDCFGQSGLGWTNW*ECYLGIGE*DEGF 1407
D S+R D G D* SE QGD +S LG + W*+ +G E*D G
Sbjct: 181 DPE**CASQRQPYGRDFTGED**SKSEDIERQGDTSCESRLGQSEW*KLDVGA*E*DGGV 360
Query: 1408 VSGVVSFR* 1416
+S VV R*
Sbjct: 361 LSRVVCMR* 387
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 60.5 bits (145), Expect = 5e-09
Identities = 27/63 (42%), Positives = 35/63 (54%), Gaps = 4/63 (6%)
Frame = +1
Query: 235 CYRCGTLGHYANDCKKDISCHKCGKAGHKAAEC----KDVARDVTCYNCGEKGHISTKCT 290
C+ C GH A+ C + CH CGKAGH+A EC K C NC ++GHI+ +CT
Sbjct: 538 CWNCKEPGHMASSCPNEGICHTCGKAGHRARECTVPQKPPGDLRLCNNCYKQGHIAVECT 717
Query: 291 KPK 293
K
Sbjct: 718 NEK 726
Score = 56.6 bits (135), Expect = 7e-08
Identities = 28/79 (35%), Positives = 39/79 (48%)
Frame = +1
Query: 211 PYNKDKRKKREVGGGSKPSLADIKCYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDV 270
PY +D R+ G S+ +L C C GHY +C CH C GH A+EC
Sbjct: 379 PYRRDSRR-----GFSQDNL----CKNCKRPGHYVRECPNVAVCHNCSLPGHIASECSTK 531
Query: 271 ARDVTCYNCGEKGHISTKC 289
+ C+NC E GH+++ C
Sbjct: 532 S---LCWNCKEPGHMASSC 579
Score = 55.5 bits (132), Expect = 1e-07
Identities = 23/69 (33%), Positives = 36/69 (51%)
Frame = +1
Query: 235 CYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTKCTKPKK 294
C+ C GH A++C C C + GH A+ C + + C+ CG+ GH + +CT P+K
Sbjct: 481 CHNCSLPGHIASECSTKSLCWNCKEPGHMASSCPN---EGICHTCGKAGHRARECTVPQK 651
Query: 295 AAGKVFALN 303
G + N
Sbjct: 652 PPGDLRLCN 678
Score = 48.9 bits (115), Expect = 1e-05
Identities = 27/86 (31%), Positives = 40/86 (46%), Gaps = 1/86 (1%)
Frame = +1
Query: 207 GVGKPYNKDKRKKREVGGGSKPSLADIK-CYRCGTLGHYANDCKKDISCHKCGKAGHKAA 265
G+ K + RE KP D++ C C GH A +C + +C+ C K GH A
Sbjct: 589 GICHTCGKAGHRARECTVPQKPP-GDLRLCNNCYKQGHIAVECTNEKACNNCRKTGHLAR 765
Query: 266 ECKDVARDVTCYNCGEKGHISTKCTK 291
+C + D C C GH++ +C K
Sbjct: 766 DCPN---DPICNLCNISGHVARQCPK 834
Score = 44.3 bits (103), Expect = 3e-04
Identities = 23/75 (30%), Positives = 29/75 (38%), Gaps = 20/75 (26%)
Frame = +1
Query: 235 CYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDV--------------------ARDV 274
C C GH A DC D C+ C +GH A +C RDV
Sbjct: 730 CNNCRKTGHLARDCPNDPICNLCNISGHVARQCPKSNVIGDRGGGGSLRGGYRDGGFRDV 909
Query: 275 TCYNCGEKGHISTKC 289
C +C + GH+S C
Sbjct: 910 VCRSCQQFGHMSRDC 954
Score = 37.7 bits (86), Expect = 0.032
Identities = 17/47 (36%), Positives = 21/47 (44%), Gaps = 2/47 (4%)
Frame = +1
Query: 223 GGGSKPSLADIKCYRCGTLGHYANDCKKD--ISCHKCGKAGHKAAEC 267
GG D+ C C GH + DC + C CG GH+A EC
Sbjct: 877 GGYRDGGFRDVVCRSCQQFGHMSRDCMGGPLMICQNCGGRGHQAYEC 1017
>TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA POLYMERASE II
{Encephalitozoon cuniculi}, partial (0%)
Length = 1247
Score = 57.4 bits (137), Expect = 4e-08
Identities = 27/80 (33%), Positives = 41/80 (50%), Gaps = 2/80 (2%)
Frame = -2
Query: 26 PTYNGRFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTTHMLAEEAEYWWTNAKGRLEIVG 85
P + G P+ W+Q MER+F ++QKV++ L + A WW N K R + G
Sbjct: 724 PDFEGNLQPDDLLDWLQIMERLFKYKEVLEEQKVKIVAAKLKKLASIWWENVKRRRKREG 545
Query: 86 --EVVTWAKFKAEFLRKYFP 103
++ TW K + + RKY P
Sbjct: 544 KSKIKTWEKMRQKLTRKYLP 485
>BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent virus}, partial
(1%)
Length = 726
Score = 53.5 bits (127), Expect = 6e-07
Identities = 24/78 (30%), Positives = 41/78 (51%), Gaps = 2/78 (2%)
Frame = +2
Query: 26 PTYNGRFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTTHMLAEEAEYWWTNAKGRLEIVG 85
P + G+ P+ W+Q +ER+F +++QKV++ L + A WW N K + G
Sbjct: 347 PDF*GKLQPDEFVDWLQTIERVFKYKEVAEEQKVKIVAAKLKKHASIWWKNLKRKRNCEG 526
Query: 86 --EVVTWAKFKAEFLRKY 101
++ TW K + + RKY
Sbjct: 527 KSKIKTWDKMRQKLTRKY 580
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 52.0 bits (123), Expect = 2e-06
Identities = 25/80 (31%), Positives = 40/80 (49%), Gaps = 2/80 (2%)
Frame = +2
Query: 26 PTYNGRFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTTHMLAEEAEYWWTNAKGRLEIVG 85
P + G + W+Q +ER+F ++QKV++ L + A WW N K R + G
Sbjct: 629 PDFEGNLQLDDFLDWLQTIERVFEYKEVPEEQKVKIVAAKLKKHALIWWENLKRRRKREG 808
Query: 86 --EVVTWAKFKAEFLRKYFP 103
++ TW K + + RKY P
Sbjct: 809 KSKIKTWDKMRQKLTRKYLP 868
>TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (39%)
Length = 630
Score = 51.2 bits (121), Expect = 3e-06
Identities = 26/93 (27%), Positives = 36/93 (37%)
Frame = +3
Query: 205 GHGVGKPYNKDKRKKREVGGGSKPSLADIKCYRCGTLGHYANDCKKDISCHKCGKAGHKA 264
GH D+ + + GG D CY CG+ H+A DC + + G G+
Sbjct: 372 GHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGG 551
Query: 265 AECKDVARDVTCYNCGEKGHISTKCTKPKKAAG 297
+CY CG GHI+ C P G
Sbjct: 552 G-------GTSCYRCGGVGHIARDCATPSSGGG 629
>TC82733 similar to GP|10177404|dbj|BAB10535.
gene_id:K24M7.12~pir||S42136~similar to unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 710
Score = 50.1 bits (118), Expect = 6e-06
Identities = 32/110 (29%), Positives = 45/110 (40%), Gaps = 18/110 (16%)
Frame = +3
Query: 210 KPYNKDKRKKREVGGGSKPSLADIK---------CYRCGTLGHYANDC------KKDISC 254
K NK KRKK + + ++ C+ C L H A C +K+ C
Sbjct: 180 KKKNKFKRKKPDSNSKPRTGKRPLRVPGMKPGDSCFICKGLDHIAKFCTQKAEWEKNKIC 359
Query: 255 HKCGKAGHKAAECKDVARDVTC---YNCGEKGHISTKCTKPKKAAGKVFA 301
+C + GH+A C D YNCG+ GH C P + G +FA
Sbjct: 360 LRCRRRGHRAQNCPDGGSKEDFKY*YNCGDNGHSLANCPHPLQEGGTMFA 509
Score = 44.3 bits (103), Expect = 3e-04
Identities = 26/74 (35%), Positives = 34/74 (45%), Gaps = 11/74 (14%)
Frame = +3
Query: 235 CYRCGTLGHYANDC-----KKDISC-HKCGKAGHKAAECKDVARD-----VTCYNCGEKG 283
C RC GH A +C K+D + CG GH A C ++ C+ C E+G
Sbjct: 357 CLRCRRRGHRAQNCPDGGSKEDFKY*YNCGDNGHSLANCPHPLQEGGTMFAQCFVCKEQG 536
Query: 284 HISTKCTKPKKAAG 297
H+S C PK A G
Sbjct: 537 HLSKNC--PKNAHG 572
>TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 974
Score = 47.0 bits (110), Expect = 5e-05
Identities = 29/84 (34%), Positives = 37/84 (43%), Gaps = 2/84 (2%)
Frame = +2
Query: 213 NKDKRKKRE-VGGGSKPSLADIKCYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDVA 271
++D R RE +G G P +C+ CG GH+A DCK +KC
Sbjct: 296 SRDSRDSREYLGRGPPPGSG--RCFNCGIDGHWARDCKAGDWKNKC-------------- 427
Query: 272 RDVTCYNCGEKGHISTKC-TKPKK 294
Y CGE+GHI C PKK
Sbjct: 428 -----YRCGERGHIEKNCKNSPKK 484
>TC92636 homologue to GP|15042313|gb|AAK82093.1 232R {Chilo iridescent
virus}, partial (1%)
Length = 772
Score = 35.0 bits (79), Expect(2) = 0.001
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 2/51 (3%)
Frame = +1
Query: 55 DDQKVRLTTHMLAEEAEYWWTNAKGRLEIVGE--VVTWAKFKAEFLRKYFP 103
++ KV++ L + A WW N K R + G+ + TW K + RKY P
Sbjct: 526 EEHKVKIVAVKLKKHALIWWENLKRRRKCEGKSKIKTWDKMCRKLTRKYSP 678
Score = 26.2 bits (56), Expect(2) = 0.001
Identities = 12/43 (27%), Positives = 21/43 (47%)
Frame = +3
Query: 6 HRSEGKVEDRRLDRFLRNNPPTYNGRFDPEGAQTWVQGMERIF 48
HR + + D ++D P + G P+ W+Q +ER+F
Sbjct: 396 HRRKTLMNDIKVDI------PDFEGELQPDEFVDWLQAIERVF 506
>TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.150 -
Arabidopsis thaliana, partial (17%)
Length = 378
Score = 37.4 bits (85), Expect = 0.042
Identities = 22/68 (32%), Positives = 27/68 (39%)
Frame = +1
Query: 222 VGGGSKPSLADIKCYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDVARDVTCYNCGE 281
VGGG CY CG GH A +C G+ G +CY+CGE
Sbjct: 4 VGGG---------CYNCGESGHMARECTSG-GGGGGGRYGGGGGGGGGGGGGGSCYSCGE 153
Query: 282 KGHISTKC 289
GH + C
Sbjct: 154 SGHFARDC 177
Score = 32.3 bits (72), Expect = 1.3
Identities = 10/15 (66%), Positives = 13/15 (86%)
Frame = +1
Query: 276 CYNCGEKGHISTKCT 290
CYNCGE GH++ +CT
Sbjct: 16 CYNCGESGHMARECT 60
>AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing factor
[imported] - Arabidopsis thaliana, partial (62%)
Length = 508
Score = 36.6 bits (83), Expect = 0.071
Identities = 29/92 (31%), Positives = 42/92 (45%), Gaps = 2/92 (2%)
Frame = +2
Query: 159 RPDIYQYMCVQEIRDFDTLVHKCRKFD--DAGRVKANYYKAQSEKRGKGHGVGKPYNKDK 216
RP Y ++ + RD + R+ D + RV+ ++ S+ G G G G
Sbjct: 143 RPPGYAFIDFDDRRD---ALDAIRELDGKNGWRVELSH---NSKTGGGGGGRGGGGGGGG 304
Query: 217 RKKREVGGGSKPSLADIKCYRCGTLGHYANDC 248
+ R GGGS D+KCY CG GH+A C
Sbjct: 305 GRGRSGGGGS-----DLKCYXCGEPGHFARXC 385
Score = 29.6 bits (65), Expect = 8.7
Identities = 10/25 (40%), Positives = 13/25 (52%)
Frame = +2
Query: 273 DVTCYNCGEKGHISTKCTKPKKAAG 297
D+ CY CGE GH + C +G
Sbjct: 335 DLKCYXCGEPGHFARXCNSSPGGSG 409
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 35.4 bits (80), Expect = 0.16
Identities = 14/27 (51%), Positives = 18/27 (65%)
Frame = +3
Query: 223 GGGSKPSLADIKCYRCGTLGHYANDCK 249
GGGS D+KCY CG GH+A +C+
Sbjct: 315 GGGS-----DLKCYECGEPGHFARECR 380
Score = 30.0 bits (66), Expect = 6.7
Identities = 9/19 (47%), Positives = 15/19 (78%)
Frame = +3
Query: 251 DISCHKCGKAGHKAAECKD 269
D+ C++CG+ GH A EC++
Sbjct: 327 DLKCYECGEPGHFARECRN 383
Score = 29.6 bits (65), Expect = 8.7
Identities = 9/17 (52%), Positives = 12/17 (69%)
Frame = +3
Query: 273 DVTCYNCGEKGHISTKC 289
D+ CY CGE GH + +C
Sbjct: 327 DLKCYECGEPGHFAREC 377
>BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.5 [imported]
- Arabidopsis thaliana, partial (5%)
Length = 627
Score = 35.0 bits (79), Expect = 0.21
Identities = 15/56 (26%), Positives = 25/56 (43%)
Frame = -1
Query: 234 KCYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTKC 289
KCY+C GH+A++C + ++ A+ CY C + GH + C
Sbjct: 504 KCYKCQQPGHWASNCPSMSAANRVSGGSGGASG--------NCYKCNQPGHWANNC 361
>BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (54%)
Length = 364
Score = 33.9 bits (76), Expect = 0.46
Identities = 25/91 (27%), Positives = 39/91 (42%)
Frame = +1
Query: 159 RPDIYQYMCVQEIRDFDTLVHKCRKFDDAGRVKANYYKAQSEKRGKGHGVGKPYNKDKRK 218
RP Y ++ + RD +H+ + RV+ ++ + K G G G G
Sbjct: 145 RPPGYAFIEFDDPRDALDAIHELDG-KNGWRVQLSH----NSKSGGGGGRG--------- 282
Query: 219 KREVGGGSKPSLADIKCYRCGTLGHYANDCK 249
GG D+KCY CG GH+A +C+
Sbjct: 283 ----GGRGGRGGDDLKCYECGEPGHFARECR 363
Score = 30.0 bits (66), Expect = 6.7
Identities = 12/32 (37%), Positives = 17/32 (52%)
Frame = +1
Query: 258 GKAGHKAAECKDVARDVTCYNCGEKGHISTKC 289
G+ G + D D+ CY CGE GH + +C
Sbjct: 274 GRGGGRGGRGGD---DLKCYECGEPGHFAREC 360
Score = 29.6 bits (65), Expect = 8.7
Identities = 9/18 (50%), Positives = 14/18 (77%)
Frame = +1
Query: 251 DISCHKCGKAGHKAAECK 268
D+ C++CG+ GH A EC+
Sbjct: 310 DLKCYECGEPGHFARECR 363
>TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (16%)
Length = 421
Score = 33.9 bits (76), Expect = 0.46
Identities = 17/48 (35%), Positives = 22/48 (45%)
Frame = +3
Query: 211 PYNKDKRKKREVGGGSKPSLADIKCYRCGTLGHYANDCKKDISCHKCG 258
PY +D + G S+ +L C C GHYA +C CH CG
Sbjct: 303 PYRRDSSR-----GFSRDNL----CKNCKRPGHYARECPNVAVCHNCG 419
Score = 33.1 bits (74), Expect = 0.79
Identities = 14/31 (45%), Positives = 18/31 (57%)
Frame = +3
Query: 250 KDISCHKCGKAGHKAAECKDVARDVTCYNCG 280
+D C C + GH A EC +VA C+NCG
Sbjct: 336 RDNLCKNCKRPGHYARECPNVA---VCHNCG 419
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.350 0.157 0.564
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 49,084,501
Number of Sequences: 36976
Number of extensions: 780669
Number of successful extensions: 6820
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 2619
Number of HSP's successfully gapped in prelim test: 329
Number of HSP's that attempted gapping in prelim test: 3910
Number of HSP's gapped (non-prelim): 3340
length of query: 1419
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1311
effective length of database: 5,021,319
effective search space: 6582949209
effective search space used: 6582949209
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC144724.7


