

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144724.10 - phase: 0 /pseudo
(1449 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
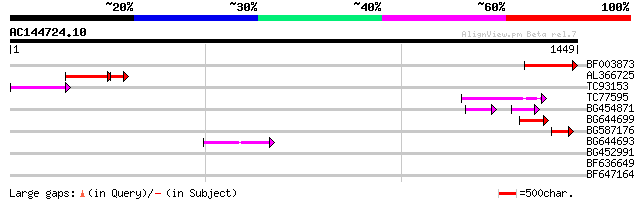
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 257 3e-68
AL366725 119 1e-35
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 91 4e-18
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 90 6e-18
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 72 1e-16
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 65 2e-10
BG587176 weakly similar to PIR|G84493|G84 probable retroelement ... 55 2e-07
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 53 1e-06
BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, pa... 37 0.056
BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativ... 31 4.0
BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis t... 30 8.9
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 257 bits (656), Expect = 3e-68
Identities = 128/135 (94%), Positives = 131/135 (96%)
Frame = +2
Query: 1315 GVGRALKSKKLTPRFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSHV 1374
GVGRALKSKKLT RFIGPYQISERVGTVAYRVGLPPHL NLHDVFHVSQLRKYVPDPSHV
Sbjct: 2 GVGRALKSKKLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLRKYVPDPSHV 181
Query: 1375 IQSDDVQVRDNLTVETLPLRIDDRKVKTLRGKEIPLVRVVWDGATGESLTWELESKMLES 1434
IQSDDVQVRDNLTVETLP+RIDDRKVKTLRGKEIPLVRVVWD A GESLTWELESKM+ES
Sbjct: 182 IQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMVES 361
Query: 1435 YPKLFA*GKFSRTKI 1449
YP+LFA*GKFSRTKI
Sbjct: 362 YPELFA*GKFSRTKI 406
>AL366725
Length = 485
Score = 119 bits (297), Expect(2) = 1e-35
Identities = 77/120 (64%), Positives = 84/120 (69%)
Frame = +3
Query: 142 IRIKLQRLLSFRSVSSSRMDCGLR*RGQLDTNRSEFSPSW*TVAGFMRRIPRLITRL*MS 201
IR L RLLS R+ SSSRM G RGQ DTN SEFS * A RRI RL+TRL*MS
Sbjct: 9 IRTMLLRLLSSRNASSSRMV*GPTSRGQSDTNSSEFSLI**IPAESTRRILRLMTRL*MS 188
Query: 202 GRLRANKVALSPIVPLLIRANRGWSRIDDQGRRMLLLRLSVLIVARKTTRAMLVMER*RD 261
GR RAN+VALS IVPLLIRANR WS I RRMLL RLSV +ARK TR M V++R R+
Sbjct: 189 GRPRANRVALSLIVPLLIRANREWSMIGVLRRRMLLRRLSVSTMARKATRVMFVLKRSRN 368
Score = 51.2 bits (121), Expect(2) = 1e-35
Identities = 29/44 (65%), Positives = 33/44 (74%)
Frame = +1
Query: 259 *RDKEMFPMW*EGSCTG*LQL**HCVLQLQ*RGSHWFSMQAT*E 302
*RD+EM P+ EGS *LQ *+CVLQLQ RGS+WF MQA *E
Sbjct: 352 *RDQEMCPV*QEGSYCS*LQAK*YCVLQLQRRGSYWFPMQAA*E 483
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 90.5 bits (223), Expect = 4e-18
Identities = 66/155 (42%), Positives = 91/155 (58%)
Frame = +3
Query: 1 LKLWGNSLMQMLVRMLRRGCLRPL*GIILLLSREDITPMEPRRGLRRSTEFSVLCNALKF 60
LKL+ NS +++++ ++ GC R *GII SR + M R G RR E+S CN L+
Sbjct: 51 LKLF-NSYLRLILVVMEPGC*RLS*GIIHQHSRVGMLLMVLRSG*RRLKEYSESCNVLRH 227
Query: 61 RRCDLVHISWLRKLMMGGLVFYLP*SKMVLL*LGLCLGGSS*ADTFRKMFAGRRRLSSLS 120
RRC L L++ M+GGLV L S+M+L * G C G S A F++M R+RL+ S
Sbjct: 228 RRCSLGRTC*LKRPMIGGLVCCLFWSRMMLW*PGPCSGRSFWAGIFQRMSGVRKRLNFWS 407
Query: 121 *SKKTCL*LSMLPSLWSWQIFIRIKLQRLLSFRSV 155
*++ TCL SML SLW+ I ++R LS SV
Sbjct: 408 *NRVTCLSQSMLQSLWNLPHSTLITVRRQLSSPSV 512
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 90.1 bits (222), Expect = 6e-18
Identities = 64/222 (28%), Positives = 104/222 (46%), Gaps = 5/222 (2%)
Frame = +2
Query: 1155 LSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTW 1214
+SDR + RFW+ G LS++YHPQTDG +ER Q ++ +LR V W
Sbjct: 581 VSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGTERWNQEIQAVLRAYVCWSQDNW 760
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
LP ++ N ++SSIG PF +G ++GG V G Q +++K
Sbjct: 761 GDLLPTVQLALRNRHNSSIGATPFFVEHGYHVDPIPTVEDTGGVVSEGEAAAQLLVKRMK 940
Query: 1275 ----MIQEKMKASQSRQKSYHDKRRKDLE-FQEGDHVFLRVTPVTGVGRALKSKKLTPRF 1329
IQ ++ A+Q R ++ +KRR + +Q GD V+L V+ ++ + K
Sbjct: 941 DVTGFIQAEIVAAQQRSEASANKRRCPADRYQVGDKVWLNVSNY----KSPRPSKKLDWL 1108
Query: 1330 IGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDP 1371
Y+++ V + +P ++ FHV LR+ DP
Sbjct: 1109 HHKYEVTRFVTPHVVELNVP---GTVYPKFHVDLLRRAASDP 1225
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 72.4 bits (176), Expect(2) = 1e-16
Identities = 37/81 (45%), Positives = 46/81 (56%)
Frame = +2
Query: 1164 SRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEF 1223
S FWK L + G+ L +SSAYHP +DGQSE + E LR + W P E+
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 1224 TYNNSYHSSIGMAPFEALYGR 1244
YN SY+ S M PF+ALYGR
Sbjct: 212 WYNTSYNISAAMTPFKALYGR 274
Score = 33.5 bits (75), Expect(2) = 1e-16
Identities = 22/73 (30%), Positives = 34/73 (46%), Gaps = 2/73 (2%)
Frame = +1
Query: 1283 SQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALK--SKKLTPRFIGPYQISERVG 1340
+Q K DK+R+ EFQ G+HV +++ P AL+ K +P F +
Sbjct: 388 AQQTMKHQADKKRRHFEFQLGEHVLVKLQPYQQSSVALRKYQKFGSPNFGSLLTVCSL*V 567
Query: 1341 TVAYRVGLPPHLS 1353
A+ PP+LS
Sbjct: 568 ESAFHCKSPPYLS 606
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 65.1 bits (157), Expect = 2e-10
Identities = 32/74 (43%), Positives = 49/74 (65%), Gaps = 1/74 (1%)
Frame = +2
Query: 1304 DHVFLRVTPVT-GVGRALKSKKLTPRFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVS 1362
+ V L+V P G R K KL+ R+IGP+++ +R+G VAY + LPP LS +H VFHVS
Sbjct: 2 EQVLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVS 181
Query: 1363 QLRKYVPDPSHVIQ 1376
++Y D +++I+
Sbjct: 182 MFKRYHGDGNYIIR 223
>BG587176 weakly similar to PIR|G84493|G84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 729
Score = 55.5 bits (132), Expect = 2e-07
Identities = 25/54 (46%), Positives = 36/54 (66%)
Frame = -1
Query: 1386 LTVETLPLRIDDRKVKTLRGKEIPLVRVVWDGATGESLTWELESKMLESYPKLF 1439
L +ET P+RI DR K +R K I +V++VWD + E +TWE E++M YP+ F
Sbjct: 717 LDLETRPVRILDRMEKAMRKKPIQMVKIVWDCSGREEITWETEARMKADYPEWF 556
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 52.8 bits (125), Expect = 1e-06
Identities = 55/185 (29%), Positives = 92/185 (49%), Gaps = 3/185 (1%)
Frame = +1
Query: 495 ASRERS*VFY*SYSWNGAGVDGTLSYISFRVS*VEETVRRFA*EEVC*TKCFTLGSAGFV 554
+SR +*++Y* + + ++ LS S ++ VE +RF + T ++G G V
Sbjct: 19 SSRMEN*LWY*FVTKYESYLNSFLSDKSIKIESVEAPTQRFTRKGFHST*YLSIGCCGLV 198
Query: 555 SEEERW*YEVVYRLSSVEQSNYQE*VSTSEN**FDGSVSGCTHF---QQD*FEIRLSPD* 611
EEERW + LS + +++ VS+ * ++ F QD* ++ ++P *
Sbjct: 199 FEEERWVP*NEH*LSPTK*RQHKDKVSSPSY*---RTI**SPRF*MVFQD*LKVGITPT* 369
Query: 612 SKR*RYAEDSFQNAVWSL*I*SYAFRCYECTWSVYGVHESHLSCILGSIRGCFYL*YFNL 671
R*R + + F N +WSL* S F + +YG +E + I CF *YF++
Sbjct: 370 GNR*RCS*NCFPN*IWSL*NPSDVFWVNKSPDGIYGTNE*SFPRLPRLISDCF***YFDI 549
Query: 672 LQD*R 676
LQ+*+
Sbjct: 550 LQE*K 564
>BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, partial (33%)
Length = 560
Score = 37.0 bits (84), Expect = 0.056
Identities = 26/59 (44%), Positives = 33/59 (55%)
Frame = +2
Query: 935 VRVA*TV*RYELSV*VVTSECETGNAED***ISEEYQRST*S*CQVCRLVGC**SD*RW 993
+ + *TV R E +* V +ECE G+ ED I YQR + C+V R *SD*RW
Sbjct: 59 LELP*TVQRLEFGL*SVATECEIGDVEDQQRIFG*YQRGSEGGCEVGRFNVWE*SD*RW 235
>BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativa}, partial
(4%)
Length = 653
Score = 30.8 bits (68), Expect = 4.0
Identities = 35/166 (21%), Positives = 65/166 (39%), Gaps = 4/166 (2%)
Frame = +1
Query: 1188 TDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCR 1247
+D Q+ +LE L EQ G + L E YN ++H + G PF+ +Y
Sbjct: 7 SDEQAGLLNHTLETHLLYFTSEQQGV*NFFLTWAECLYNTNFHRTAGCTPFKVVY---VV 177
Query: 1248 TQLCWFESGGRVVLGPEIVQQTTEKVKMIQEKMKASQSRQKSY----HDKRRKDLEFQEG 1303
L F ++ E + +++ + + +Y H +R + +
Sbjct: 178 AHLQKFVVARDLIYRNEGLHKSST*TSFGRGTRAYEALSRPAYETC*HPRRPLSIVYTRD 357
Query: 1304 DHVFLRVTPVTGVGRALKSKKLTPRFIGPYQISERVGTVAYRVGLP 1349
+V P K GPYQ+ +++G+VA+++ LP
Sbjct: 358 RTYEWQVLP-----------KYVA*CYGPYQVIKQIGSVAFKL*LP 462
>BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana},
partial (6%)
Length = 469
Score = 29.6 bits (65), Expect = 8.9
Identities = 21/69 (30%), Positives = 33/69 (47%)
Frame = +1
Query: 1278 EKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQISE 1337
E K + R K +HD+R EF+EG+ V L + + L KL + GP+Q+
Sbjct: 142 ENAKIYKERTKKWHDRRIIRREFREGELVLLFNSRL-----KLFPGKLRSHWSGPFQVKN 306
Query: 1338 RVGTVAYRV 1346
+ + A V
Sbjct: 307 VMPSGAVEV 333
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.362 0.162 0.603
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,486,165
Number of Sequences: 36976
Number of extensions: 724756
Number of successful extensions: 7300
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 2173
Number of HSP's successfully gapped in prelim test: 276
Number of HSP's that attempted gapping in prelim test: 4975
Number of HSP's gapped (non-prelim): 2770
length of query: 1449
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1341
effective length of database: 5,021,319
effective search space: 6733588779
effective search space used: 6733588779
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (22.0 bits)
S2: 65 (29.6 bits)
Medicago: description of AC144724.10


