

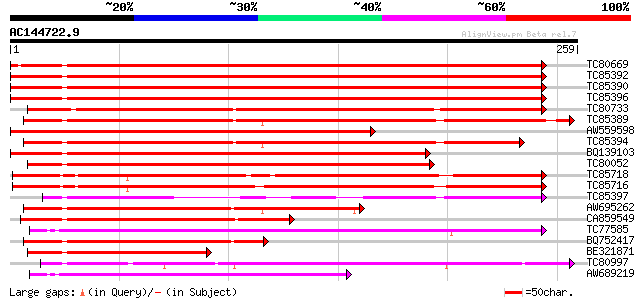
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.9 + phase: 0
(259 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 355 1e-98
TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chape... 353 5e-98
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 352 6e-98
TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protei... 351 2e-97
TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa prot... 312 7e-86
TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protei... 295 2e-80
AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 ... 284 2e-77
TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic retic... 276 4e-75
BQ139103 homologue to GP|2655420|gb| heat shock cognate protein ... 275 1e-74
TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chapero... 244 2e-65
TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa prot... 231 2e-61
TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chape... 229 1e-60
TC85397 homologue to GP|170386|gb|AAA99920.1|| glucose-regulated... 178 2e-45
AW695262 homologue to GP|170386|gb|A glucose-regulated protein 7... 167 4e-42
CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus sto... 161 2e-40
TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock... 154 2e-38
BQ752417 similar to GP|5597020|dbj ER chaperone BiP {Aspergillus... 130 4e-31
BE321871 GP|20563125|db heat shock cognate protein {Bombyx mori}... 120 7e-28
TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [im... 117 3e-27
AW689219 similar to PIR|D96830|D96 probable heat-shock protein ... 103 9e-23
>TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (44%)
Length = 955
Score = 355 bits (910), Expect = 1e-98
Identities = 175/245 (71%), Positives = 208/245 (84%)
Frame = +2
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 98 MATK-EGKAIGIDLGTTYSCVGVW--QNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAA 268
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NPQNTVFDAKRLIGR+FSD VQ D+ WPFKV+ G +KPMI V YKG+EK
Sbjct: 269 KNQVAMNPQNTVFDAKRLIGRRFSDESVQNDMKLWPFKVVPGPAEKPMIVVNYKGEEKKF 448
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L M+++AEA+L PVKNAVVTVPAYFNDSQR+AT +AGAI+GLNV+RIIN
Sbjct: 449 AAEEISSMVLIKMREVAEAFLGHPVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIIN 628
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 629 EPTAAAIAYGLDKKASRKGEQNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 808
Query: 241 FDNRM 245
FDNRM
Sbjct: 809 FDNRM 823
>TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chaperone
hsc70-3 - tomato, complete
Length = 2295
Score = 353 bits (905), Expect = 5e-98
Identities = 175/245 (71%), Positives = 204/245 (82%)
Frame = +1
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 130 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 303
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+ SD VQ D+ WPFKVI G DKPMI V YK +EK
Sbjct: 304 KNQVAMNPTNTVFDAKRLIGRRISDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQF 483
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L MK+IAEAYL + +KNAVVTVPAYFNDSQR+AT +AG I+GLNVMRIIN
Sbjct: 484 SAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 663
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 664 EPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 843
Query: 241 FDNRM 245
FDNRM
Sbjct: 844 FDNRM 858
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 352 bits (904), Expect = 6e-98
Identities = 174/245 (71%), Positives = 204/245 (83%)
Frame = +1
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 136 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 309
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+F D VQ D+ WPFKVI G DKPMI V YK +EK
Sbjct: 310 KNQVAMNPTNTVFDAKRLIGRRFGDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQF 489
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L MK+IAEAYL + +KNAVVTVPAYFNDSQR+AT +AG I+GLNV+RIIN
Sbjct: 490 SAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVLRIIN 669
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 670 EPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 849
Query: 241 FDNRM 245
FDNRM
Sbjct: 850 FDNRM 864
>TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protein 70
{Cucumis sativus}, complete
Length = 2320
Score = 351 bits (900), Expect = 2e-97
Identities = 173/245 (70%), Positives = 205/245 (83%)
Frame = +2
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 107 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 280
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+ SD VQ D+ WPFKVI+G +KPMI V YKG+EK
Sbjct: 281 KNQVAMNPTNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEEKLF 460
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
+EEISS++L M++IAEAYL +KNAVVTVPAYFNDSQR+AT +AG IAGLNV+RIIN
Sbjct: 461 ASEEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 640
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 641 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 820
Query: 241 FDNRM 245
FDNRM
Sbjct: 821 FDNRM 835
>TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa protein
(HSP70). {Neurospora crassa}, partial (54%)
Length = 1258
Score = 312 bits (800), Expect = 7e-86
Identities = 158/237 (66%), Positives = 192/237 (80%)
Frame = +2
Query: 9 AVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINP 68
A+GIDLGTTYSCVG + +K +EII ND+GN+ TPS+VAF D +RL+G AAK+Q A+NP
Sbjct: 188 AIGIDLGTTYSCVGRYANDK--IEIIANDQGNRTTPSYVAFNDTERLIGDAAKNQVAMNP 361
Query: 69 QNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSI 128
NTVFDAKRLIGRKFSD VQ D+ +PFKVI KP I V++KG+ K EEIS++
Sbjct: 362 HNTVFDAKRLIGRKFSDSEVQADMKHFPFKVIDK-GGKPNIEVEFKGENKTFTPEEISAM 538
Query: 129 ILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAAIA 188
+L M++ AEAYL V NAV+TVPAYFNDSQR+AT +AG IAGLNV+RIIN+PTAAAIA
Sbjct: 539 VLVKMRETAEAYLGGQVTNAVITVPAYFNDSQRQATKDAGLIAGLNVLRIINEPTAAAIA 718
Query: 189 YGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
YGLD K +GERN+ +FDLGGGTFDVSLLTI+ +FEVK+TAG+THLGGEDFDNR+
Sbjct: 719 YGLDKK--AEGERNVLIFDLGGGTFDVSLLTIEEGIFEVKSTAGDTHLGGEDFDNRL 883
>TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protein 4
precursor (BiP 4) (78 kDa glucose-regulated protein
homolog 4) (GRP 78-4)., partial (94%)
Length = 2265
Score = 295 bits (754), Expect = 2e-80
Identities = 155/253 (61%), Positives = 197/253 (77%), Gaps = 1/253 (0%)
Frame = +2
Query: 7 GHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAI 66
G +GIDLGTTYSCVGV+ + VEII ND+GN+ITPS+V+FTDD+RL+G AAK+ AA+
Sbjct: 155 GTVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 328
Query: 67 NPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYK-GQEKNLCAEEI 125
NP+ T+FD KRLIGRKF+D VQ+D+ P+K+++ + KP I V+ K G+ K EE+
Sbjct: 329 NPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIVNK-DGKPYIQVRVKDGETKVFSPEEV 505
Query: 126 SSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAA 185
S++IL MK+ AEA+L +++AVVTVPAYFND+QR+AT +AG IAGLNV RIIN+PTAA
Sbjct: 506 SAMILTKMKETAEAFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 685
Query: 186 AIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
AIAYGLD KG GE+NI VFDLGGGTFDVS+LTI VFEV +T G+THLGGEDFD R+
Sbjct: 686 AIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLSTNGDTHLGGEDFDQRI 856
Query: 246 CWNKMCFTMNLIK 258
M + + LIK
Sbjct: 857 ----MEYFIKLIK 883
>AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 kDa protein.
[Petunia] {Petunia hybrida}, partial (24%)
Length = 532
Score = 284 bits (727), Expect = 2e-77
Identities = 138/167 (82%), Positives = 155/167 (92%)
Frame = +1
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA + +AVGIDLGTTYSCV VWLEEKNRVEIIHNDEG+KITPSFVAFTD QRLVG AA
Sbjct: 31 MANNCECYAVGIDLGTTYSCVAVWLEEKNRVEIIHNDEGSKITPSFVAFTDGQRLVGAAA 210
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDI+ WPFKV+SG+NDKPMI++K+KGQEK L
Sbjct: 211 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDILLWPFKVVSGVNDKPMISLKFKGQEKLL 390
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNA 167
CAEEISSI+L NM++IAE YL+SPVKNA +TVPAYFND+QRKAT++A
Sbjct: 391 CAEEISSIVLTNMREIAEMYLESPVKNAGITVPAYFNDAQRKATIDA 531
>TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic reticulum
HSC70-cognate binding protein precursor {Glycine max},
partial (38%)
Length = 962
Score = 276 bits (707), Expect = 4e-75
Identities = 144/230 (62%), Positives = 180/230 (77%), Gaps = 1/230 (0%)
Frame = +1
Query: 7 GHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAI 66
G +GIDLGTTYSCVGV+ + VEII ND+GN+ITPS+V+FTDD+RL+G AAK+ AA+
Sbjct: 289 GTVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 462
Query: 67 NPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYK-GQEKNLCAEEI 125
NP+ T+FD KRLIGRKF D VQ+D+ P+K+++ + KP I V+ K G+ K EEI
Sbjct: 463 NPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNR-DGKPYIQVRVKDGETKVFSPEEI 639
Query: 126 SSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAA 185
S++IL MK+ AE +L +++AVVTVPAYFND+QR+AT +AG IAGLNV RIIN+PTAA
Sbjct: 640 SAMILGKMKETAEGFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 819
Query: 186 AIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTH 235
AIAYGLD KG GE+NI VFDLGGGTFDVS+LTI VFEV AT G+TH
Sbjct: 820 AIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTH 960
>BQ139103 homologue to GP|2655420|gb| heat shock cognate protein HSC70
{Brassica napus}, partial (29%)
Length = 628
Score = 275 bits (703), Expect = 1e-74
Identities = 136/192 (70%), Positives = 162/192 (83%)
Frame = +2
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 56 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 229
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ D+ WPFK+ISG +KP+I V YKG++K
Sbjct: 230 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKIISGPAEKPLIGVNYKGEDKEF 409
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L M++IAEAYL S +KNAVVTVPAYFNDSQR+AT +AG IAGLNVMRIIN
Sbjct: 410 AAEEISSMVLMKMREIAEAYLGSAIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 589
Query: 181 DPTAAAIAYGLD 192
+PTAAAIAYGLD
Sbjct: 590 EPTAAAIAYGLD 625
>TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chaperone hsc70-3
- tomato, partial (27%)
Length = 667
Score = 244 bits (624), Expect = 2e-65
Identities = 121/186 (65%), Positives = 148/186 (79%)
Frame = +3
Query: 9 AVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINP 68
A+GIDLGTTYSCV V + ++II ND G + TPSFVAF D +R++G AA + AA NP
Sbjct: 105 AIGIDLGTTYSCVAVC--KNGEIDIIVNDLGKRTTPSFVAFKDSERMIGDAAFNIAASNP 278
Query: 69 QNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSI 128
NT+FDAKRLIGRKFSDP+VQ D+ WPFKVI LNDKPMI V Y +EK+ AEEISS+
Sbjct: 279 TNTIFDAKRLIGRKFSDPIVQSDVKLWPFKVIGDLNDKPMIVVNYNDEEKHFAAEEISSM 458
Query: 129 ILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAAIA 188
+L M++IAE +L S V++ V+TVPAYFNDSQR++T +AGAIAGLNVMRIIN+PTAAAIA
Sbjct: 459 VLVKMREIAETFLGSIVEDVVITVPAYFNDSQRQSTRDAGAIAGLNVMRIINEPTAAAIA 638
Query: 189 YGLDNK 194
YG + K
Sbjct: 639 YGFNTK 656
>TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa protein
mitochondrial precursor. [Kidney bean French bean]
{Phaseolus vulgaris}, complete
Length = 2371
Score = 231 bits (590), Expect = 2e-61
Identities = 130/245 (53%), Positives = 167/245 (68%), Gaps = 1/245 (0%)
Frame = +2
Query: 2 ARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDD-QRLVGGAA 60
+R +GIDLGTT SCV V +E KN +++ N EG + TPS VAFT + LVG A
Sbjct: 239 SRAAGNDVIGIDLGTTNSCVSV-MEGKNP-KVVENSEGARTTPSVVAFTQKGELLVGTPA 412
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K QA NP+NT+ AKRLIGR+F DP QK++ P+K++ N V+ KGQ+ +
Sbjct: 413 KRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAW--VEAKGQQYS- 583
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
+I + +L MK+ AEAYL V AV+TVPAYFND+QR+AT +AG IAGL V+RIIN
Sbjct: 584 -PSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIIN 760
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAA++YG++ +G I VFDLGGGTFDVS+L I VFEVKAT G+T LGGED
Sbjct: 761 EPTAAALSYGMNKEGL------IAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGED 922
Query: 241 FDNRM 245
FDN +
Sbjct: 923 FDNAL 937
>TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chaperone PHSP1
precursor mitochondrial - garden pea, partial (98%)
Length = 2422
Score = 229 bits (583), Expect = 1e-60
Identities = 127/245 (51%), Positives = 162/245 (65%), Gaps = 1/245 (0%)
Frame = +1
Query: 2 ARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDD-QRLVGGAA 60
+R +GIDLGTT SCV + +E KN ++I N EG + TPS VAF + LVG A
Sbjct: 223 SRPAGSDVIGIDLGTTNSCVSL-MEGKNP-KVIENSEGARTTPSVVAFNQKGELLVGTPA 396
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K QA NP NT+F KRLIGR+F DP QK++ P+K++ N + + ++
Sbjct: 397 KRQAVTNPTNTLFGTKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEIN----KQQY 564
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
+I + +L MK+ AEAYL + AVVTVPAYFND+QR+AT +AG IAGL V RIIN
Sbjct: 565 SPSQIGAFVLTKMKETAEAYLGKTISKAVVTVPAYFNDAQRQATKDAGRIAGLEVKRIIN 744
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAA++YG++NK E I VFDLGGGTFDVS+L I VFEVKAT G+T LGGED
Sbjct: 745 EPTAAALSYGMNNK-----EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGED 909
Query: 241 FDNRM 245
FDN +
Sbjct: 910 FDNAL 924
>TC85397 homologue to GP|170386|gb|AAA99920.1|| glucose-regulated protein 78
{Lycopersicon esculentum}, partial (57%)
Length = 648
Score = 178 bits (451), Expect = 2e-45
Identities = 108/230 (46%), Positives = 136/230 (58%)
Frame = +1
Query: 16 TTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINPQNTVFDA 75
TTYSCVGV+ + VEII ND+GN+ITPS+V+FTDD+RL+G AAK+ AA+NP+ T+FD
Sbjct: 1 TTYSCVGVY--KNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAVNPERTIFDV 174
Query: 76 KRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSIILANMKK 135
K +I K++ +E + +MK
Sbjct: 175 ------------------------------KRLIGRKFEDKE-----------VQRDMKL 231
Query: 136 IAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAAIAYGLDNKG 195
+ + K + +AT +AG IAGLNV RIIN+PTAAAIAYGLD KG
Sbjct: 232 VPYKIVNRDGKPYI------------QATKDAGVIAGLNVARIINEPTAAAIAYGLDKKG 375
Query: 196 RCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
GE+NI VFDLGGGTFDVS+LTI VFEV AT G+THLGGEDFD R+
Sbjct: 376 ---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRI 516
>AW695262 homologue to GP|170386|gb|A glucose-regulated protein 78
{Lycopersicon esculentum}, partial (39%)
Length = 607
Score = 167 bits (423), Expect = 4e-42
Identities = 87/158 (55%), Positives = 120/158 (75%), Gaps = 2/158 (1%)
Frame = +2
Query: 7 GHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAI 66
G +GIDLGTTYSCVGV+ + VEII ND+GN+ITPS+V+FTDD+RL+G AAK+ AA+
Sbjct: 128 GTVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 301
Query: 67 NPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYK-GQEKNLCAEEI 125
NP+ T+FD KRLIGRKF+D VQ+D+ P+K+++ + KP I V+ K G+ K EE+
Sbjct: 302 NPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIVN-KDGKPYIQVRVKDGETKVFSPEEV 478
Query: 126 SSIILANMKKIAEAYLKSPVKNAVVTVPAYF-NDSQRK 162
S++IL MK+ AEA+L +++AVVTVPA ND+QR+
Sbjct: 479 SAMILTKMKETAEAFLGKTIRDAVVTVPALLSNDAQRQ 592
>CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus stolonifer},
partial (19%)
Length = 527
Score = 161 bits (408), Expect = 2e-40
Identities = 81/125 (64%), Positives = 98/125 (77%)
Frame = +3
Query: 6 DGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAA 65
+G AVGIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AAK+Q A
Sbjct: 156 EGKAVGIDLGTTYSCVGVW--QNDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVA 329
Query: 66 INPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEI 125
+NP NTVFDAKRLIGR+F+D VQ D+ WPFKVI KP I V+YKG+ K EEI
Sbjct: 330 MNPYNTVFDAKRLIGRRFADQEVQSDMKHWPFKVIDKA-AKPYIQVEYKGETKQFTPEEI 506
Query: 126 SSIIL 130
SS++L
Sbjct: 507 SSMVL 521
>TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock protein
{Arabidopsis thaliana}, partial (92%)
Length = 3027
Score = 154 bits (390), Expect = 2e-38
Identities = 80/237 (33%), Positives = 136/237 (56%), Gaps = 1/237 (0%)
Frame = +1
Query: 10 VGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINPQ 69
VG D G SC+ V + + ++++ NDE + TP+ V F D QR +G A +NP+
Sbjct: 247 VGFDFGNE-SCI-VAVARQRGIDVVLNDESKRETPAIVCFGDKQRFIGTAGAASTMMNPK 420
Query: 70 NTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSII 129
N++ KRLIG+KF+DP +Q+D+ PF V G + P+I +Y G+ + A ++ ++
Sbjct: 421 NSISQIKRLIGKKFADPELQRDLKSLPFNVTEGPDGYPLIHARYLGESREFTATQVFGMM 600
Query: 130 LANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAAIAY 189
L+N+K+IA+ L + V + + +P YF D QR++ ++A IAGL+ + +I++ TA A+AY
Sbjct: 601 LSNLKEIAQKNLNAAVVDCCIGIPVYFTDLQRRSVLDAATIAGLHPLHLIHETTATALAY 780
Query: 190 GLDNKGRCDGE-RNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
G+ + E N+ D+G + V + K V + + + LGG DFD +
Sbjct: 781 GIYKTDLPENEWLNVAFVDVGHASMQVCIAGFKKGQLHVLSHSYDRSLGGRDFDEAL 951
>BQ752417 similar to GP|5597020|dbj ER chaperone BiP {Aspergillus oryzae},
partial (18%)
Length = 690
Score = 130 bits (328), Expect = 4e-31
Identities = 64/112 (57%), Positives = 86/112 (76%)
Frame = +1
Query: 7 GHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAI 66
G +GIDLGTTYSCVGV + +VEI+ ND+GN+ITPS+VAFT+++RLVG AAK+QAA
Sbjct: 322 GTVIGIDLGTTYSCVGVM--QGGKVEILVNDQGNRITPSYVAFTEEERLVGDAAKNQAAA 495
Query: 67 NPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEK 118
NP NT+FD KR+IG+KF+D VQ D+ +P+KVI + KP++ V +G K
Sbjct: 496 NPYNTIFDIKRMIGQKFADKAVQSDVKHFPYKVIE-KDGKPVVEVDVQGSAK 648
>BE321871 GP|20563125|db heat shock cognate protein {Bombyx mori}, partial
(13%)
Length = 346
Score = 120 bits (300), Expect = 7e-28
Identities = 58/84 (69%), Positives = 70/84 (83%)
Frame = +1
Query: 9 AVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINP 68
AVGIDLGTTYSCVGV+ + +VEII ND+GN+ TPS+VAFTD +RL+G AAK+Q A+NP
Sbjct: 94 AVGIDLGTTYSCVGVF--QHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMNP 267
Query: 69 QNTVFDAKRLIGRKFSDPVVQKDI 92
NT+FDAKRLIGRKF D VQ D+
Sbjct: 268 NNTIFDAKRLIGRKFEDATVQADM 339
>TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [imported] -
Arabidopsis thaliana, partial (91%)
Length = 1717
Score = 117 bits (294), Expect = 3e-27
Identities = 81/255 (31%), Positives = 131/255 (50%), Gaps = 11/255 (4%)
Frame = +3
Query: 15 GTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINPQ----N 70
GT+ V VW + VE++ N K+ SFV F D+ G Q + + +
Sbjct: 3 GTSQCSVAVW--NGSEVELLKNKRNQKLMKSFVTFKDEAP--SGGVTSQFSHEHEMLFGD 170
Query: 71 TVFDAKRLIGRKFSDPVVQKDIMFWPFKVIS-GLNDKPMIAVKYKGQEKNLCAEEISSII 129
T+F+ KRLIGR +DPVV PF V + + +P IA ++ EE+ ++
Sbjct: 171 TIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAMF 347
Query: 130 LANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAAIAY 189
L ++ + E +LK P++N V+TVP F+ Q A A+AGL+V+R++ +PTA A+ Y
Sbjct: 348 LVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALLY 527
Query: 190 GLDNKGRCD------GERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDN 243
G + E+ +F++G G DV++ G V ++KA AG+T +GGED
Sbjct: 528 GQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDLLQ 704
Query: 244 RMCWNKMCFTMNLIK 258
M + + + N+ K
Sbjct: 705 NMMRHLLPDSENIFK 749
>AW689219 similar to PIR|D96830|D96 probable heat-shock protein 41956-44878
[imported] - Arabidopsis thaliana, partial (20%)
Length = 534
Score = 103 bits (256), Expect = 9e-23
Identities = 51/147 (34%), Positives = 85/147 (57%)
Frame = +3
Query: 10 VGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINPQ 69
VG D G SC+ V + + ++++ NDE + TP+ V F + QR +G A +NP+
Sbjct: 87 VGFDFGNE-SCI-VAVARQRGIDVVLNDESKRETPAIVCFGEKQRFIGTAGAASTMMNPK 260
Query: 70 NTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSII 129
N++ KRLIG+ +SDP +Q+D+ PF V G + P+I +Y G+ K ++ +++
Sbjct: 261 NSISQIKRLIGKLYSDPDLQRDLKSLPFSVAEGPDGYPLIQARYLGEVKMFTPTQVFAMM 440
Query: 130 LANMKKIAEAYLKSPVKNAVVTVPAYF 156
L NMK+IAE L + + + +P YF
Sbjct: 441 LGNMKEIAEKNLNAAXNDCCIGIPCYF 521
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,036,158
Number of Sequences: 36976
Number of extensions: 79456
Number of successful extensions: 459
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 421
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 422
length of query: 259
length of database: 9,014,727
effective HSP length: 94
effective length of query: 165
effective length of database: 5,538,983
effective search space: 913932195
effective search space used: 913932195
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC144722.9


