

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.19 + phase: 0
(628 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
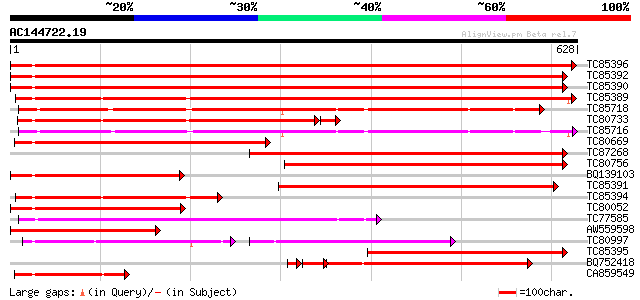
Score E
Sequences producing significant alignments: (bits) Value
TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protei... 781 0.0
TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chape... 781 0.0
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 780 0.0
TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protei... 615 e-176
TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa prot... 454 e-128
TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa prot... 427 e-123
TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chape... 430 e-121
TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 421 e-118
TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chape... 392 e-109
TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 332 2e-91
BQ139103 homologue to GP|2655420|gb| heat shock cognate protein ... 286 2e-77
TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chape... 282 2e-76
TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic retic... 273 1e-73
TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chapero... 251 8e-67
TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock... 245 3e-65
AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 ... 236 3e-62
TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [im... 145 9e-60
TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protei... 221 7e-58
BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated pr... 194 8e-56
CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus sto... 171 1e-42
>TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protein 70
{Cucumis sativus}, complete
Length = 2320
Score = 781 bits (2018), Expect = 0.0
Identities = 399/630 (63%), Positives = 502/630 (79%), Gaps = 3/630 (0%)
Frame = +2
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVWQ +DRVEII NDQGNRTTPS VAFT+S+RLIGDAA
Sbjct: 107 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 280
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
KNQ A NP+NT+FD KRLIGR+ SD+ VQ D LWPFKVI+G KP I V Y G+EK F
Sbjct: 281 KNQVAMNPTNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEEKLF 460
Query: 121 VAEEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIIN 180
+EEISS++L KMREIAEA+L +KNAV+TVPAYFNDSQR+ATKDAG IAGLNV+RIIN
Sbjct: 461 ASEEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 640
Query: 181 EPTAAALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGED 240
EPTAAA+AYGL K+A V ++N+ IFDLGGGTFDVS+LT+++ +EVKATAGDTHLGGED
Sbjct: 641 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 820
Query: 241 FDNRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID 300
FDNRM+THFV++FK+KN +DIS NP+ALRRLRTACERAKRTLS + TIE+D++++G+D
Sbjct: 821 FDNRMVTHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVD 1000
Query: 301 FSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQE 360
F ++ITRA+FEE+NM+LF KCME V CL DAKM+K++V DVVLVGGS+RIPKV+QLLQ+
Sbjct: 1001FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQD 1180
Query: 361 FFQGKELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSV 419
FF GKELC SINPDEAVAYGAA+Q A+L E + V +L+L DVTPLS G+ G +M+V
Sbjct: 1181FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSQGLETAGGVMTV 1360
Query: 420 VIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHP 478
+IPRNT+IP K+ + T D+Q VLI+VYEGER + DNNLLG F LS +P APRG P
Sbjct: 1361LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTKDNNLLGKFELSGIPPAPRGVP 1540
Query: 479 H-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKF 537
VCF IDA+GILNVSA ++T+G KN+ITITN+KGRLS+ +IE+M+QEAE +K++D +
Sbjct: 1541QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEH 1720
Query: 538 KKKVEARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVF 597
KKKVEA+N+L++Y Y +R +K++ SSKL+ DK++I A+ + +Q + F
Sbjct: 1721KKKVEAKNSLENYAYNMRNTIKDEKISSKLSGGDKKQIEDAIEGAIQWLDANQLAEADEF 1900
Query: 598 VDFLKELESIFESAMNKINKGDSDEKSDSD 627
D +KELE+I + K+ +G + E + D
Sbjct: 1901EDKMKELETICNPIIAKMYQGGAGEGPEVD 1990
>TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chaperone
hsc70-3 - tomato, complete
Length = 2295
Score = 781 bits (2017), Expect = 0.0
Identities = 397/621 (63%), Positives = 497/621 (79%), Gaps = 3/621 (0%)
Frame = +1
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVWQ +DRVEII NDQGNRTTPS VAFT+++RLIGDAA
Sbjct: 130 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 303
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
KNQ A NP+NT+FD KRLIGR+ SD VQ D LWPFKVI G +KP I+V Y +EK+F
Sbjct: 304 KNQVAMNPTNTVFDAKRLIGRRISDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQF 483
Query: 121 VAEEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIIN 180
AEEISS++L KM+EIAEA+L + +KNAV+TVPAYFNDSQR+ATKDAG I+GLNVMRIIN
Sbjct: 484 SAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 663
Query: 181 EPTAAALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGED 240
EPTAAA+AYGL K+A ++N+ IFDLGGGTFDVS+LT+++ +EVKATAGDTHLGGED
Sbjct: 664 EPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 843
Query: 241 FDNRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID 300
FDNRM+ HFV++FK+KN +DIS NP+ALRR+RTACERAKRTLS + TIE+D++++GID
Sbjct: 844 FDNRMVNHFVQEFKRKNKKDISGNPRALRRVRTACERAKRTLSSTAQTTIEIDSLFEGID 1023
Query: 301 FSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQE 360
F ++ITRA+FEE+NM+LF KCME V CL DAKM+KNSVDDVVLVGGS+RIPKV+QLLQ+
Sbjct: 1024FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVDDVVLVGGSTRIPKVQQLLQD 1203
Query: 361 FFQGKELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSV 419
FF GKELC SINPDEAVAYGAA+Q A+L E + V +L+L DVTPLSLG+ G +M+V
Sbjct: 1204FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 1383
Query: 420 VIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHP 478
+IPRNT+IP K+ + T D+Q VLI+VYEGER + DNNLLG F LS +P APRG P
Sbjct: 1384LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVP 1563
Query: 479 H-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKF 537
VCF IDA+GILNVSA ++T+G KN+ITITN+KGRLS+ IE+M+QEAE +K++D +
Sbjct: 1564QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEH 1743
Query: 538 KKKVEARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVF 597
K+KVEA+NAL++Y Y +R +K+D +SKL+ DK+KI A+ + +Q + F
Sbjct: 1744KRKVEAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEADEF 1923
Query: 598 VDFLKELESIFESAMNKINKG 618
D +KELE I + ++ +G
Sbjct: 1924EDKMKELEGICNPIIARMYQG 1986
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 780 bits (2015), Expect = 0.0
Identities = 396/621 (63%), Positives = 497/621 (79%), Gaps = 3/621 (0%)
Frame = +1
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVWQ +DRVEII NDQGNRTTPS VAFT+++RLIGDAA
Sbjct: 136 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 309
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
KNQ A NP+NT+FD KRLIGR++ D VQ D LWPFKVI G +KP I+V Y +EK+F
Sbjct: 310 KNQVAMNPTNTVFDAKRLIGRRFGDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQF 489
Query: 121 VAEEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIIN 180
AEEISS++L KM+EIAEA+L + +KNAV+TVPAYFNDSQR+ATKDAG I+GLNV+RIIN
Sbjct: 490 SAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVLRIIN 669
Query: 181 EPTAAALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGED 240
EPTAAA+AYGL K+A ++N+ IFDLGGGTFDVS+LT+++ +EVKATAGDTHLGGED
Sbjct: 670 EPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 849
Query: 241 FDNRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID 300
FDNRM+ HFV++FK+KN +DIS NP+ALRRLRTACERAKRTLS + TIE+D++++GID
Sbjct: 850 FDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGID 1029
Query: 301 FSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQE 360
F ++ITRA+FEE+NM+LF KCME V CL DAKM+KNSV DVVLVGGS+RIPKV+QLLQ+
Sbjct: 1030FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQD 1209
Query: 361 FFQGKELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSV 419
FF GKELC SINPDEAVAYGAA+Q A+L E + V +L+L DVTPLSLG+ G +M+V
Sbjct: 1210FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 1389
Query: 420 VIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHP 478
+IPRNT+IP K+ + T D+Q VLI+VYEGER + DNNLLG F LS +P APRG P
Sbjct: 1390LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVP 1569
Query: 479 H-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKF 537
VCF IDA+GILNVSA ++T+G KN+ITITN+KGRLS+ +IE+M+QEAE +K++D +
Sbjct: 1570QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEH 1749
Query: 538 KKKVEARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVF 597
KKKVEA+NAL++Y Y +R +K+D +SKL+ DK+KI A+ + +Q + F
Sbjct: 1750KKKVEAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEADEF 1929
Query: 598 VDFLKELESIFESAMNKINKG 618
D +KELE I + ++ +G
Sbjct: 1930EDKMKELEGICNPIIARMYQG 1992
>TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protein 4
precursor (BiP 4) (78 kDa glucose-regulated protein
homolog 4) (GRP 78-4)., partial (94%)
Length = 2265
Score = 615 bits (1586), Expect = e-176
Identities = 325/633 (51%), Positives = 454/633 (71%), Gaps = 12/633 (1%)
Frame = +2
Query: 7 GVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDLGTTYSCVGV++ N VEII NDQGNR TPS V+FT+ +RLIG+AAKN AA
Sbjct: 155 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 328
Query: 67 NPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYM-GKEKRFVAEEI 125
NP TIFDVKRLIGRK++D VQ D L P+K+++ + KP I V+ G+ K F EE+
Sbjct: 329 NPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIVN-KDGKPYIQVRVKDGETKVFSPEEV 505
Query: 126 SSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAA 185
S++IL+KM+E AEAFL +++AV+TVPAYFND+QR+ATKDAG IAGLNV RIINEPTAA
Sbjct: 506 SAMILTKMKETAEAFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 685
Query: 186 ALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRM 245
A+AYGL K+ ++NI +FDLGGGTFDVSILT+ + +EV +T GDTHLGGEDFD R+
Sbjct: 686 AIAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLSTNGDTHLGGEDFDQRI 856
Query: 246 LTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSI 305
+ +F+K KKK+++DIS++ +AL +LR ERAKR LS + +E+++++ G+DFS +
Sbjct: 857 MEYFIKLIKKKHSKDISKDNRALGKLRRESERAKRALSSQHQVRVEIESLFDGVDFSEPL 1036
Query: 306 TRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGK 365
TRA+FEE+N +LF K M V + DA ++KN +D++VLVGGS+RIPKV+QLL+++F GK
Sbjct: 1037TRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFDGK 1216
Query: 366 ELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRN 424
E +NPDEAVA+GAA+Q ++L E + +++L DV PL+LG+ G +M+ +IPRN
Sbjct: 1217EPNKGVNPDEAVAFGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRN 1396
Query: 425 TSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHPH-KVC 482
T IP K++ + T +D Q+TV I+V+EGER D LLG F LS +P APRG P +V
Sbjct: 1397TVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGNFDLSGIPPAPRGTPQIEVT 1576
Query: 483 FTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVE 542
F +DA+GILNV A ++ +G +ITITNEKGRLS+ +I+RM++EAE F +D K K++++
Sbjct: 1577FEVDANGILNVRAEDKGTGKSEKITITNEKGRLSQEEIDRMVREAEEFAEEDKKVKERID 1756
Query: 543 ARNALDDYLYKVR-KVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDFL 601
ARNAL+ Y+Y ++ ++ D + KL +KEKI +A+ + + D+Q + F + L
Sbjct: 1757ARNALETYVYNMKNQISDKDKLADKLESDEKEKIEAAVKEALEWLDDNQTVEKEEFEEKL 1936
Query: 602 KELESIFESAMNKINK-------GDSDEKSDSD 627
KE+E++ + + + G S E D D
Sbjct: 1937KEVEAVCNPIITAVYQRSGGAPGGASGEGEDED 2035
>TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa protein
mitochondrial precursor. [Kidney bean French bean]
{Phaseolus vulgaris}, complete
Length = 2371
Score = 454 bits (1167), Expect = e-128
Identities = 266/590 (45%), Positives = 368/590 (62%), Gaps = 7/590 (1%)
Frame = +2
Query: 10 IGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNS-QRLIGDAAKNQAATNP 68
IGIDLGTT SCV V + +N +V + N +G RTTPS VAFT + L+G AK QA TNP
Sbjct: 263 IGIDLGTTNSCVSVMEGKNPKV--VENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNP 436
Query: 69 SNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEISSV 128
NTI KRLIGR++ D Q + + P+K++ N + K +++ +I +
Sbjct: 437 ENTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWV----EAKGQQYSPSQIGAF 604
Query: 129 ILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAAALA 188
+L+KM+E AEA+L V AVITVPAYFND+QR+ATKDAG IAGL V+RIINEPTAAAL+
Sbjct: 605 VLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALS 784
Query: 189 YGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRMLTH 248
YG+ K I +FDLGGGTFDVSIL + + +EVKAT GDT LGGEDFDN +L
Sbjct: 785 YGMNKEG------LIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDF 946
Query: 249 FVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID----FSSS 304
V +FK+ + D+S++ AL+RLR A E+AK LS ++ I L I + +
Sbjct: 947 LVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNIT 1126
Query: 305 ITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQG 364
+TR+KFE + NL E+ SCL DA + +D+V+LVGG +R+PKV++++ F G
Sbjct: 1127LTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-G 1303
Query: 365 KELCNSINPDEAVAYGAAIQAVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRN 424
K C +NPDEAVA GAA+Q + VK L+L DVTPLSLG+ G I + +I RN
Sbjct: 1304KSPCKGVNPDEAVAMGAALQGGILRGDVK---ELLLLDVTPLSLGIETLGGIFTRLINRN 1474
Query: 425 TSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSL-SLPLAPRGHPH-KVC 482
T+IP K++ + T D+Q+ V IKV +GER SDN +LG F L LP APRG P +V
Sbjct: 1475TTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVT 1654
Query: 483 FTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVE 542
F IDA+GI+ VSA ++++G + +ITI G LS +I+ M++EAE +D + K ++
Sbjct: 1655FDIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLID 1831
Query: 543 ARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQE 592
RN+ D +Y + K + ++ V KE ++ ++ GDS E
Sbjct: 1832IRNSADTSIYSIEKSL-SEYREKIPAEVAKEIEDAVSDLRSAMAGDSADE 1978
>TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa protein
(HSP70). {Neurospora crassa}, partial (54%)
Length = 1258
Score = 427 bits (1098), Expect(2) = e-123
Identities = 216/335 (64%), Positives = 272/335 (80%)
Frame = +2
Query: 9 AIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNP 68
AIGIDLGTTYSCVG + ND++EII NDQGNRTTPS VAF +++RLIGDAAKNQ A NP
Sbjct: 188 AIGIDLGTTYSCVGRYA--NDKIEIIANDQGNRTTPSYVAFNDTERLIGDAAKNQVAMNP 361
Query: 69 SNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEISSV 128
NT+FD KRLIGRK+SDS VQ D +PFKVI KP I V++ G+ K F EEIS++
Sbjct: 362 HNTVFDAKRLIGRKFSDSEVQADMKHFPFKVID-KGGKPNIEVEFKGENKTFTPEEISAM 538
Query: 129 ILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAAALA 188
+L KMRE AEA+L V NAVITVPAYFNDSQR+ATKDAG IAGLNV+RIINEPTAAA+A
Sbjct: 539 VLVKMRETAEAYLGGQVTNAVITVPAYFNDSQRQATKDAGLIAGLNVLRIINEPTAAAIA 718
Query: 189 YGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRMLTH 248
YGL K+A +RN+ IFDLGGGTFDVS+LT+++ +EVK+TAGDTHLGGEDFDNR++ H
Sbjct: 719 YGLDKKAE--GERNVLIFDLGGGTFDVSLLTIEEGIFEVKSTAGDTHLGGEDFDNRLVNH 892
Query: 249 FVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSITRA 308
FV +FK+K+ +D+S N +ALRRLRTACERAKRTLS + +IE+D++++GIDF +SITRA
Sbjct: 893 FVNEFKRKHKKDLSSNARALRRLRTACERAKRTLSSSAQTSIEIDSLFEGIDFYTSITRA 1072
Query: 309 KFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVV 343
+FEE+ +LF ++ V+ L+DAK++K+ V ++V
Sbjct: 1073RFEELCQDLFRSTIQPVDRVLSDAKIDKSQVHEIV 1177
Score = 34.3 bits (77), Expect(2) = e-123
Identities = 12/22 (54%), Positives = 20/22 (90%)
Frame = +3
Query: 345 VGGSSRIPKVRQLLQEFFQGKE 366
VGGS+RIP++++L+ ++F GKE
Sbjct: 1182 VGGSTRIPRIQKLISDYFNGKE 1247
>TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chaperone PHSP1
precursor mitochondrial - garden pea, partial (98%)
Length = 2422
Score = 430 bits (1106), Expect = e-121
Identities = 265/632 (41%), Positives = 378/632 (58%), Gaps = 13/632 (2%)
Frame = +1
Query: 10 IGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNS-QRLIGDAAKNQAATNP 68
IGIDLGTT SCV + + +N +V I N +G RTTPS VAF + L+G AK QA TNP
Sbjct: 247 IGIDLGTTNSCVSLMEGKNPKV--IENSEGARTTPSVVAFNQKGELLVGTPAKRQAVTNP 420
Query: 69 SNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEISSV 128
+NT+F KRLIGR++ D Q + + P+K++ N + + ++++ +I +
Sbjct: 421 TNTLFGTKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEIN----KQQYSPSQIGAF 588
Query: 129 ILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAAALA 188
+L+KM+E AEA+L + AV+TVPAYFND+QR+ATKDAG IAGL V RIINEPTAAAL+
Sbjct: 589 VLTKMKETAEAYLGKTISKAVVTVPAYFNDAQRQATKDAGRIAGLEVKRIINEPTAAALS 768
Query: 189 YGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRMLTH 248
YG+ + I +FDLGGGTFDVSIL + + +EVKAT GDT LGGEDFDN +L
Sbjct: 769 YGMNNKEGL-----IAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDF 933
Query: 249 FVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID----FSSS 304
V +FK+ ++ D++++ AL+RLR A E+AK LS ++ I L I + +
Sbjct: 934 LVSEFKRTDSIDLAKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNIT 1113
Query: 305 ITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQG 364
+TR+KFE + NL E+ SCL DA + VD+V+LVGG +R+PKV++++ E F G
Sbjct: 1114LTRSKFEALVNNLIERTKAPCKSCLKDANISIKDVDEVLLVGGMTRVPKVQEVVSEIF-G 1290
Query: 365 KELCNSINPDEAVAYGAAIQAVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRN 424
K +NPDEAVA GAA+Q + VK L+L DVTPLSLG+ G I + +I RN
Sbjct: 1291KSPSKGVNPDEAVAMGAALQGGILRGDVK---ELLLLDVTPLSLGIETLGGIFTRLISRN 1461
Query: 425 TSIPVKRTNEYVTTEDDQST--VLIKVYEGERLKDSDNNLLGLFSL-SLPLAPRGHPH-K 480
T+IP K++ + T D+Q+ EG R +DN LG F L +P APRG P +
Sbjct: 1462TTIPTKKSQVFSTAADNQTQRGYXRCSQEGXREMAADNKSLGEFDLVGIPPAPRGLPQIE 1641
Query: 481 VCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKK 540
V F IDA+GI+ VSA ++++G + +ITI G LS +I M++EAE +D + K
Sbjct: 1642VTFDIDANGIVTVSAKDKSTGKEQQITI-RSSGGLSDDEINNMVKEAELHAQRDQERKAL 1818
Query: 541 VEARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDF 600
++ +N+ D +Y + K + ++ + V KE NS ++ G+S VD
Sbjct: 1819IDIKNSADTSIYSIEKSL-SEYREKIPSEVAKEIENSVSDLRTAMEGES--------VDE 1971
Query: 601 LKELESIFESAMNKINK----GDSDEKSDSDS 628
+K A++KI + G S SD S
Sbjct: 1972IKTKLDAANKAVSKIGQHMSGGSSGGSSDGGS 2067
>TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (44%)
Length = 955
Score = 421 bits (1082), Expect = e-118
Identities = 207/284 (72%), Positives = 242/284 (84%)
Frame = +2
Query: 6 EGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAA 65
EG AIGIDLGTTYSCVGVWQ NDRVEII NDQGNRTTPS VAFT+++RLIGDAAKNQ A
Sbjct: 110 EGKAIGIDLGTTYSCVGVWQ--NDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKNQVA 283
Query: 66 TNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEI 125
NP NT+FD KRLIGR++SD VQ D LWPFKV+ G KP I+V Y G+EK+F AEEI
Sbjct: 284 MNPQNTVFDAKRLIGRRFSDESVQNDMKLWPFKVVPGPAEKPMIVVNYKGEEKKFAAEEI 463
Query: 126 SSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAA 185
SS++L KMRE+AEAFL PVKNAV+TVPAYFNDSQR+ATKDAG I+GLNV+RIINEPTAA
Sbjct: 464 SSMVLIKMREVAEAFLGHPVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAA 643
Query: 186 ALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRM 245
A+AYGL K+A+ ++N+ IFDLGGGTFDVS+LT+++ +EVKATAGDTHLGGEDFDNRM
Sbjct: 644 AIAYGLDKKASRKGEQNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRM 823
Query: 246 LTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEAT 289
+ HFV +F++KN +DIS N +ALRRLRTACERAKRTLS + T
Sbjct: 824 VNHFVSEFRRKNKKDISGNARALRRLRTACERAKRTLSSTAQTT 955
>TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chaperone
HSC71.0 - garden pea, partial (59%)
Length = 1336
Score = 392 bits (1007), Expect = e-109
Identities = 204/356 (57%), Positives = 266/356 (74%), Gaps = 3/356 (0%)
Frame = +2
Query: 266 KALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSITRAKFEEMNMNLFEKCMETV 325
+ALRRLRTACERAKRTLS + TIE+D++++GIDF S ITRA+FEE+NM+LF KCME V
Sbjct: 2 RALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFYSPITRARFEELNMDLFRKCMEPV 181
Query: 326 NSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGKELCNSINPDEAVAYGAAIQ- 384
CL DAKM+K SV DVVLVGGS+RIPKV+QLLQ+FF GKELC SINPDEAVAYGAA+Q
Sbjct: 182 EKCLRDAKMDKKSVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQA 361
Query: 385 AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRNTSIPVKRTNEYVTTEDDQST 444
A+L E + V +L+L DVTPLSLG+ G +M+V+IPRNT+IP K+ + T D+Q
Sbjct: 362 AILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPG 541
Query: 445 VLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHPH-KVCFTIDADGILNVSAVEETSGN 502
VLI+V+EGER + DNNLLG F LS +P APRG P VCF IDA+GILNVSA ++T+G
Sbjct: 542 VLIQVFEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQ 721
Query: 503 KNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVEARNALDDYLYKVRKVMKNDS 562
KN+ITITN+KGRLS+ IE+M+QEAE +K++D + KKKVEA+NAL++Y Y +R +K++
Sbjct: 722 KNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTIKDEK 901
Query: 563 TSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDFLKELESIFESAMNKINKG 618
+ KL DK+KI + + +Q + F D +KELE + + K+ +G
Sbjct: 902 IAGKLDSDDKKKIEDTIEAAIQWLDANQLAEADEFEDKMKELEGVCNPIIAKMYQG 1069
>TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (53%)
Length = 1189
Score = 332 bits (852), Expect = 2e-91
Identities = 173/317 (54%), Positives = 233/317 (72%), Gaps = 3/317 (0%)
Frame = +3
Query: 305 ITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQG 364
ITRA+FEE+NM+LF KCME V CL DAK++K+ V +VVLVGGS+RIPKV+QLLQ+FF G
Sbjct: 3 ITRARFEELNMDLFRKCMEPVEKCLRDAKIDKSHVHEVVLVGGSTRIPKVQQLLQDFFNG 182
Query: 365 KELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPR 423
KELC SINPDEAVAYGAA+Q A+L E + V +L+L DVTPLSLG+ G +M+ +IPR
Sbjct: 183 KELCKSINPDEAVAYGAAVQAAILSGEGDEKVQDLLLLDVTPLSLGLETAGGVMTTLIPR 362
Query: 424 NTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHPH-KV 481
NT+IP K+ + T D+Q VLI+V+EGER + DNNLLG F L+ +P APRG P V
Sbjct: 363 NTTIPTKKEQIFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELTGIPPAPRGVPQINV 542
Query: 482 CFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKV 541
CF IDA+GILNVSA ++T+G KN+ITITN+KGRLS+ +IE+M+++AE +KA+D + KKKV
Sbjct: 543 CFDIDANGILNVSAEDKTAGVKNKITITNDKGRLSKEEIEKMVKDAEKYKAEDEEVKKKV 722
Query: 542 EARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDFL 601
EA+N++++Y Y +R +K++ KL+ DKEKI A+ + +Q + F D
Sbjct: 723 EAKNSIENYAYNMRNTIKDEKIGGKLSHEDKEKIEKAVEDAIQWLEGNQMAEVDEFEDKQ 902
Query: 602 KELESIFESAMNKINKG 618
KELE I + K+ +G
Sbjct: 903 KELEGICNPIIAKMYQG 953
>BQ139103 homologue to GP|2655420|gb| heat shock cognate protein HSC70
{Brassica napus}, partial (29%)
Length = 628
Score = 286 bits (732), Expect = 2e-77
Identities = 146/193 (75%), Positives = 163/193 (83%)
Frame = +2
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVWQ +DRVEII NDQGNRTTPS VAFT+S+RLIGDAA
Sbjct: 56 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 229
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
KNQ A NP NT+FD KRLIGR++SD+ VQ D LWPFK+ISG KP I V Y G++K F
Sbjct: 230 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKIISGPAEKPLIGVNYKGEDKEF 409
Query: 121 VAEEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIIN 180
AEEISS++L KMREIAEA+L S +KNAV+TVPAYFNDSQR+ATKDAG IAGLNVMRIIN
Sbjct: 410 AAEEISSMVLMKMREIAEAYLGSAIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 589
Query: 181 EPTAAALAYGLQK 193
EPTAAA+AYGL K
Sbjct: 590 EPTAAAIAYGLDK 628
>TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chaperone BiP-A
- soybean, partial (52%)
Length = 1231
Score = 282 bits (722), Expect = 2e-76
Identities = 147/314 (46%), Positives = 218/314 (68%), Gaps = 4/314 (1%)
Frame = +1
Query: 298 GIDFSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQL 357
G+ FS +TRA+FEE+N +LF K M V + DA ++KN +D++VLVGGS+RIPKV+QL
Sbjct: 4 GVAFSEPLTRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQL 183
Query: 358 LQEFFQGKELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDI 416
L+++F GKE +NPDEAVA+GAA+Q ++L E +++L DV PL+LG+ G +
Sbjct: 184 LKDYFDGKEPNKGVNPDEAVAFGAAVQGSILSGEGGAETKDILLLDVAPLTLGIETVGGV 363
Query: 417 MSVVIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPR 475
M+ +IPRNT IP K++ + T +D Q+TV I+V+EGER D LLG F LS +P APR
Sbjct: 364 MTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAPR 543
Query: 476 GHPH-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQD 534
G P +V F +DA+GILNV A ++ +G +ITITNEKGRLS+ +IERM++EAE F +D
Sbjct: 544 GTPQIEVTFEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIERMVREAEEFAEED 723
Query: 535 MKFKKKVEARNALDDYLYKVR-KVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQED 593
K K++++ARNAL+ Y+Y ++ +V D + KL +KEKI +A+ + + D+Q +
Sbjct: 724 KKVKERIDARNALETYVYNMKNQVSDKDKLADKLESDEKEKIETAVKEALEWLDDNQSVE 903
Query: 594 TFVFVDFLKELESI 607
+ + LKE+E++
Sbjct: 904 KEEYEEKLKEVEAV 945
>TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic reticulum
HSC70-cognate binding protein precursor {Glycine max},
partial (38%)
Length = 962
Score = 273 bits (699), Expect = 1e-73
Identities = 146/230 (63%), Positives = 177/230 (76%), Gaps = 1/230 (0%)
Frame = +1
Query: 7 GVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDLGTTYSCVGV++ N VEII NDQGNR TPS V+FT+ +RLIG+AAKN AA
Sbjct: 289 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 462
Query: 67 NPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYM-GKEKRFVAEEI 125
NP TIFDVKRLIGRK+ D VQ D L P+K+++ + KP I V+ G+ K F EEI
Sbjct: 463 NPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVN-RDGKPYIQVRVKDGETKVFSPEEI 639
Query: 126 SSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAA 185
S++IL KM+E AE FL +++AV+TVPAYFND+QR+ATKDAG IAGLNV RIINEPTAA
Sbjct: 640 SAMILGKMKETAEGFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 819
Query: 186 ALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTH 235
A+AYGL K+ ++NI +FDLGGGTFDVSILT+ + +EV AT GDTH
Sbjct: 820 AIAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTH 960
>TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chaperone hsc70-3
- tomato, partial (27%)
Length = 667
Score = 251 bits (640), Expect = 8e-67
Identities = 131/194 (67%), Positives = 155/194 (79%)
Frame = +3
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M+ + + VAIGIDLGTTYSCV V + N ++II ND G RTTPS VAF +S+R+IGDAA
Sbjct: 81 MSGRGKKVAIGIDLGTTYSCVAVCK--NGEIDIIVNDLGKRTTPSFVAFKDSERMIGDAA 254
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
N AA+NP+NTIFD KRLIGRK+SD IVQ D LWPFKVI N+KP I+V Y +EK F
Sbjct: 255 FNIAASNPTNTIFDAKRLIGRKFSDPIVQSDVKLWPFKVIGDLNDKPMIVVNYNDEEKHF 434
Query: 121 VAEEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIIN 180
AEEISS++L KMREIAE FL S V++ VITVPAYFNDSQR++T+DAG IAGLNVMRIIN
Sbjct: 435 AAEEISSMVLVKMREIAETFLGSIVEDVVITVPAYFNDSQRQSTRDAGAIAGLNVMRIIN 614
Query: 181 EPTAAALAYGLQKR 194
EPTAAA+AYG +
Sbjct: 615 EPTAAAIAYGFNTK 656
>TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock protein
{Arabidopsis thaliana}, partial (92%)
Length = 3027
Score = 245 bits (626), Expect = 3e-65
Identities = 139/403 (34%), Positives = 223/403 (54%), Gaps = 1/403 (0%)
Frame = +1
Query: 10 IGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNPS 69
+G D G V V +++ ++++ ND+ R TP+ V F + QR IG A NP
Sbjct: 247 VGFDFGNESCIVAVARQRG--IDVVLNDESKRETPAIVCFGDKQRFIGTAGAASTMMNPK 420
Query: 70 NTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEISSVI 129
N+I +KRLIG+K++D +Q D PF V G + P I +Y+G+ + F A ++ ++
Sbjct: 421 NSISQIKRLIGKKFADPELQRDLKSLPFNVTEGPDGYPLIHARYLGESREFTATQVFGMM 600
Query: 130 LSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAAALAY 189
LS ++EIA+ L + V + I +P YF D QRR+ DA IAGL+ + +I+E TA ALAY
Sbjct: 601 LSNLKEIAQKNLNAAVVDCCIGIPVYFTDLQRRSVLDAATIAGLHPLHLIHETTATALAY 780
Query: 190 GLQKRANFVDK-RNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRMLTH 248
G+ K ++ N+ D+G + V I K V + + D LGG DFD + H
Sbjct: 781 GIYKTDLPENEWLNVAFVDVGHASMQVCIAGFKKGQLHVLSHSYDRSLGGRDFDEALFHH 960
Query: 249 FVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSITRA 308
F KFK++ D+ +N +A RLR ACE+ K+ LS + EA + ++ + D I R
Sbjct: 961 FAAKFKEEYKIDVYQNARACLRLRAACEKLKKVLSANPEAPLNIECLMDEKDVRGFIKRD 1140
Query: 309 KFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGKELC 368
FE++++ + E+ + LA+A + ++ V +VG SR+P + ++L EFF+ KE
Sbjct: 1141DFEQLSLPILERVKGPLEKALAEAGLTVENIHMVEVVGSGSRVPAINKILTEFFK-KEPR 1317
Query: 369 NSINPDEAVAYGAAIQAVLHSESVKSVPNLVLRDVTPLSLGVS 411
++N E VA GAA+Q + S + K V + + P S+ +S
Sbjct: 1318RTMNASECVARGAALQCAILSPTFK-VREFQVNESFPFSVSLS 1443
>AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 kDa protein.
[Petunia] {Petunia hybrida}, partial (24%)
Length = 532
Score = 236 bits (601), Expect = 3e-62
Identities = 114/167 (68%), Positives = 137/167 (81%)
Frame = +1
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M E A+GIDLGTTYSCV VW E+ +RVEIIHND+G++ TPS VAFT+ QRL+G AA
Sbjct: 31 MANNCECYAVGIDLGTTYSCVAVWLEEKNRVEIIHNDEGSKITPSFVAFTDGQRLVGAAA 210
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
K+QAA NP NT+FD KRLIGRK+SD +VQ D LLWPFKV+SG N+KP I +K+ G+EK
Sbjct: 211 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDILLWPFKVVSGVNDKPMISLKFKGQEKLL 390
Query: 121 VAEEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDA 167
AEEISS++L+ MREIAE +LESPVKNA ITVPAYFND+QR+AT DA
Sbjct: 391 CAEEISSIVLTNMREIAEMYLESPVKNAGITVPAYFNDAQRKATIDA 531
>TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [imported] -
Arabidopsis thaliana, partial (91%)
Length = 1717
Score = 145 bits (367), Expect(2) = 9e-60
Identities = 86/232 (37%), Positives = 129/232 (55%), Gaps = 4/232 (1%)
Frame = +1
Query: 266 KALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSITRAKFEEMNMNLFEKCMETV 325
K++ LR A + A LS + +++D + + + RA+FEE+N +FEKC +
Sbjct: 772 KSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKICKVVDRAEFEEVNKEVFEKCESLI 948
Query: 326 NSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGKELCNSINPDEAVAYGAAIQA 385
CL DAK+E ++DV+LVGG S IP+V L+ + E+ INP E GA ++
Sbjct: 949 IQCLHDAKVEVGDINDVILVGGCSYIPRVENLVTNLCKITEVYKGINPLEGAVCGATMEG 1128
Query: 386 VLHSESVKSVPNLVLRDV--TPLSLGVSIRGDIMSVVIPRNTSIPVKRTNEYVTTEDDQS 443
+ S NL L + TPL++G+ G+ VIPRNT++P ++ + T D+Q+
Sbjct: 1129 AVASGISDPFGNLDLLTIQATPLAIGIRADGNKFVPVIPRNTTMPARKDLLFTTIHDNQT 1308
Query: 444 TVLIKVYEGERLKDSDNNLLGLFS-LSLPLAPRGHPH-KVCFTIDADGILNV 493
LI VYEGE K +N+LLG F + +P AP+G P VC IDA +L V
Sbjct: 1309 EALILVYEGEGKKAEENHLLGYFKIMGIPAAPKGVPEISVCMDIDAANVLRV 1464
Score = 103 bits (257), Expect(2) = 9e-60
Identities = 75/245 (30%), Positives = 125/245 (50%), Gaps = 9/245 (3%)
Frame = +3
Query: 15 GTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAK--NQAATNPSNTI 72
GT+ V VW VE++ N + + S V F + G ++ ++ +TI
Sbjct: 3 GTSQCSVAVWN--GSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFGDTI 176
Query: 73 FDVKRLIGRKYSDSIVQMDRLLWPFKVIS-GANNKPTIIVKYMGKEKRFVAEEISSVILS 131
F++KRLIGR +D +V + L PF V + +P I + EE+ ++ L
Sbjct: 177 FNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAMFLV 353
Query: 132 KMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAAALAYGL 191
++R + E L+ P++N V+TVP F+ Q + A +AGL+V+R++ EPTA AL YG
Sbjct: 354 ELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALLYGQ 533
Query: 192 QKRANFVD------KRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRM 245
Q++ + ++ IF++G G DV++ ++KA AG T +GGED M
Sbjct: 534 QQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDLLQNM 710
Query: 246 LTHFV 250
+ H +
Sbjct: 711 MRHLL 725
>TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protein 70
{Cucumis sativus}, partial (35%)
Length = 723
Score = 221 bits (563), Expect = 7e-58
Identities = 115/224 (51%), Positives = 162/224 (71%), Gaps = 2/224 (0%)
Frame = +3
Query: 397 NLVLRDVTPLSLGVSIRGDIMSVVIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLK 456
+L+L DVTPLSLG+ G +M+V+IPRNT+IP K+ + T D+Q VLI+VYEGER +
Sbjct: 6 DLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERAR 185
Query: 457 DSDNNLLGLFSLS-LPLAPRGHPH-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGR 514
DNNLLG F LS +P APRG P VCF IDA+GILNVSA ++T+G KN+ITITN+KGR
Sbjct: 186 TRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGR 365
Query: 515 LSRGQIERMIQEAENFKAQDMKFKKKVEARNALDDYLYKVRKVMKNDSTSSKLTPVDKEK 574
LS+ +IE+M++EAE +KA+D + KKKV+A+NAL++Y Y +R +K++ SKL+P DK+K
Sbjct: 366 LSKEEIEKMVEEAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIGSKLSPEDKKK 545
Query: 575 INSAMTKGKSLIGDSQQEDTFVFVDFLKELESIFESAMNKINKG 618
I+ A+ + +Q + F D +KELES+ + K+ +G
Sbjct: 546 IDDAIDAAIQWLDSNQLAEADEFQDKMKELESLCNPIIAKMYQG 677
>BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated protein
homolog precursor (GRP 78), partial (47%)
Length = 1032
Score = 194 bits (493), Expect(3) = 8e-56
Identities = 105/233 (45%), Positives = 157/233 (67%), Gaps = 4/233 (1%)
Frame = -1
Query: 351 IPKVRQLLQEFFQGKELCNSINPDEAVAYGAAIQA-VLHSESVKSVPNLVLRDVTPLSLG 409
IPKV+ L++E+F GK+ INPDEAVA+GAA+QA VL E + +VL DV PL+LG
Sbjct: 900 IPKVQSLIEEYFGGKKASKGINPDEAVAFGAAVQAGVLSGE--EGTEEIVLMDVNPLTLG 727
Query: 410 VSIRGDIMSVVIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS 469
+ G +M+ +I RNT IP +++ + T D+Q VLI+V+EGER DNN LG F L+
Sbjct: 726 IETTGGVMTKLITRNTPIPTRKSQIFSTAADNQPVVLIQVFEGERSLTKDNNQLGKFELT 547
Query: 470 -LPLAPRGHPH-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEA 527
+P APRG P +V F +DA+GIL VSA ++ +G + ITITN+KGRL++ +I+RM++EA
Sbjct: 546 GIPPAPRGVPQIEVSFELDANGILKVSAHDKGTGKQESITITNDKGRLTKEEIDRMVEEA 367
Query: 528 ENFKAQDMKFKKKVEARNALDDYLYKVR-KVMKNDSTSSKLTPVDKEKINSAM 579
E + +D ++++EARN L++Y + ++ +V + K+ DKE I A+
Sbjct: 366 EKYAEEDKATRERIEARNGLENYAFSLKNQVNDEEGLGGKIDDEDKETILEAV 208
Score = 38.5 bits (88), Expect(3) = 8e-56
Identities = 17/29 (58%), Positives = 22/29 (75%)
Frame = -3
Query: 325 VNSCLADAKMEKNSVDDVVLVGGSSRIPK 353
V L DAK++K VDD+VLVGGS+R P+
Sbjct: 979 VEQVLKDAKVKKEDVDDIVLVGGSTRYPQ 893
Score = 24.3 bits (51), Expect(3) = 8e-56
Identities = 9/16 (56%), Positives = 14/16 (87%)
Frame = -2
Query: 308 AKFEEMNMNLFEKCME 323
AKFEE+NM+L +K ++
Sbjct: 1031 AKFEELNMDLLKKTLK 984
>CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus stolonifer},
partial (19%)
Length = 527
Score = 171 bits (432), Expect = 1e-42
Identities = 88/127 (69%), Positives = 102/127 (80%)
Frame = +3
Query: 6 EGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAA 65
EG A+GIDLGTTYSCVGVWQ NDRVEII NDQGNRTTPS VAFT+++RLIGDAAKNQ A
Sbjct: 156 EGKAVGIDLGTTYSCVGVWQ--NDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVA 329
Query: 66 TNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEI 125
NP NT+FD KRLIGR+++D VQ D WPFKVI A KP I V+Y G+ K+F EEI
Sbjct: 330 MNPYNTVFDAKRLIGRRFADQEVQSDMKHWPFKVIDKA-AKPYIQVEYKGETKQFTPEEI 506
Query: 126 SSVILSK 132
SS++L+K
Sbjct: 507 SSMVLTK 527
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.132 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,839,458
Number of Sequences: 36976
Number of extensions: 171799
Number of successful extensions: 711
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 639
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 641
length of query: 628
length of database: 9,014,727
effective HSP length: 102
effective length of query: 526
effective length of database: 5,243,175
effective search space: 2757910050
effective search space used: 2757910050
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144722.19


