

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.18 + phase: 0 /pseudo
(433 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
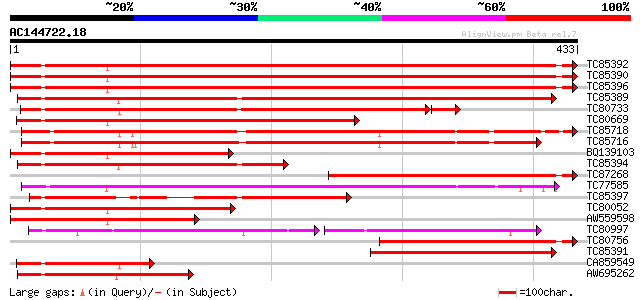
Score E
Sequences producing significant alignments: (bits) Value
TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chape... 576 e-165
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 573 e-164
TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protei... 570 e-163
TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protei... 436 e-122
TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa prot... 389 e-111
TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 382 e-106
TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa prot... 341 3e-94
TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chape... 332 2e-91
BQ139103 homologue to GP|2655420|gb| heat shock cognate protein ... 244 6e-65
TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic retic... 242 2e-64
TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chape... 229 2e-60
TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock... 216 1e-56
TC85397 homologue to GP|170386|gb|AAA99920.1|| glucose-regulated... 211 3e-55
TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chapero... 208 3e-54
AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 ... 184 6e-47
TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [im... 109 4e-45
TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 172 2e-43
TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chape... 142 3e-34
CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus sto... 133 1e-31
AW695262 homologue to GP|170386|gb|A glucose-regulated protein 7... 132 2e-31
>TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chaperone
hsc70-3 - tomato, complete
Length = 2295
Score = 576 bits (1485), Expect = e-165
Identities = 298/456 (65%), Positives = 361/456 (78%), Gaps = 23/456 (5%)
Frame = +1
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVW Q+DRVEII NDQGNRTTPS VAFT+++RLIGDAA
Sbjct: 130 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 303
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
KNQ A NP+NT+F FKVI G DKP IVVNYK EEK+F
Sbjct: 304 KNQVAMNPTNTVFDAKRLIGRRISDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQF 483
Query: 99 VAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIIN 158
AEEISS++L KM+EIAEA+L T +KNAV++VPAYFNDSQR+ATKDAG I+GLNVM+IIN
Sbjct: 484 SAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 663
Query: 159 EPTAAALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQD 218
EPTAAA+AYGL K+A ++N+ IFDLGGGTFDVS+LT++ +FEVKATAGDTHLGG+D
Sbjct: 664 EPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 843
Query: 219 FDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGID 278
FDN MV+HFV+EF+ K K+DIS N +ALRR+RTACE+AKRTLS TIE+D++++GID
Sbjct: 844 FDNRMVNHFVQEFKRKNKKDISGNPRALRRVRTACERAKRTLSSTAQTTIEIDSLFEGID 1023
Query: 279 FCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQE 338
F ++ITRA+FEE+NM+LF KCM V CL DAKMDKNSVDDVVLVGGS+RIPKV+QLLQ+
Sbjct: 1024FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVDDVVLVGGSTRIPKVQQLLQD 1203
Query: 339 VFKGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSV 397
F GKELCKSINPDEAVAYGA VQAA+LS EG++ V +L+L DVTPLS G+ G +M+V
Sbjct: 1204FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 1383
Query: 398 MIPRNTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+IPRNT+IP E+ + Y N+ V ++VY+
Sbjct: 1384LIPRNTTIPTKKEQVFSTYS----DNQPGVLIQVYE 1479
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 573 bits (1478), Expect = e-164
Identities = 297/456 (65%), Positives = 360/456 (78%), Gaps = 23/456 (5%)
Frame = +1
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVW Q+DRVEII NDQGNRTTPS VAFT+++RLIGDAA
Sbjct: 136 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 309
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
KNQ A NP+NT+F FKVI G DKP IVVNYK EEK+F
Sbjct: 310 KNQVAMNPTNTVFDAKRLIGRRFGDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQF 489
Query: 99 VAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIIN 158
AEEISS++L KM+EIAEA+L T +KNAV++VPAYFNDSQR+ATKDAG I+GLNV++IIN
Sbjct: 490 SAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVLRIIN 669
Query: 159 EPTAAALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQD 218
EPTAAA+AYGL K+A ++N+ IFDLGGGTFDVS+LT++ +FEVKATAGDTHLGG+D
Sbjct: 670 EPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 849
Query: 219 FDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGID 278
FDN MV+HFV+EF+ K K+DIS N +ALRRLRTACE+AKRTLS TIE+D++++GID
Sbjct: 850 FDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGID 1029
Query: 279 FCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQE 338
F ++ITRA+FEE+NM+LF KCM V CL DAKMDKNSV DVVLVGGS+RIPKV+QLLQ+
Sbjct: 1030FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQD 1209
Query: 339 VFKGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSV 397
F GKELCKSINPDEAVAYGA VQAA+LS EG++ V +L+L DVTPLS G+ G +M+V
Sbjct: 1210FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 1389
Query: 398 MIPRNTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+IPRNT+IP E+ + Y N+ V ++VY+
Sbjct: 1390LIPRNTTIPTKKEQVFSTYS----DNQPGVLIQVYE 1485
>TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protein 70
{Cucumis sativus}, complete
Length = 2320
Score = 570 bits (1469), Expect = e-163
Identities = 296/456 (64%), Positives = 359/456 (77%), Gaps = 23/456 (5%)
Frame = +2
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVW Q+DRVEII NDQGNRTTPS VAFT+S+RLIGDAA
Sbjct: 107 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 280
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
KNQ A NP+NT+F FKVIAG +KP I VNYKGEEK F
Sbjct: 281 KNQVAMNPTNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEEKLF 460
Query: 99 VAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIIN 158
+EEISS++L KMREIAEA+L +KNAV++VPAYFNDSQR+ATKDAG IAGLNV++IIN
Sbjct: 461 ASEEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 640
Query: 159 EPTAAALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQD 218
EPTAAA+AYGL K+A V ++N+ IFDLGGGTFDVS+LT++ +FEVKATAGDTHLGG+D
Sbjct: 641 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 820
Query: 219 FDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGID 278
FDN MV HFV+EF+ K K+DIS N +ALRRLRTACE+AKRTLS TIE+D++++G+D
Sbjct: 821 FDNRMVTHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVD 1000
Query: 279 FCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQE 338
F ++ITRA+FEE+NM+LF KCM V CL DAKMDK++V DVVLVGGS+RIPKV+QLLQ+
Sbjct: 1001FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQD 1180
Query: 339 VFKGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSV 397
F GKELCKSINPDEAVAYGA VQAA+LS EG++ V +L+L DVTPLS G+ G +M+V
Sbjct: 1181FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSQGLETAGGVMTV 1360
Query: 398 MIPRNTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+IPRNT+IP E+ + Y N+ V ++VY+
Sbjct: 1361LIPRNTTIPTKKEQVFSTYS----DNQPGVLIQVYE 1456
>TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protein 4
precursor (BiP 4) (78 kDa glucose-regulated protein
homolog 4) (GRP 78-4)., partial (94%)
Length = 2265
Score = 436 bits (1120), Expect = e-122
Identities = 228/434 (52%), Positives = 307/434 (70%), Gaps = 23/434 (5%)
Frame = +2
Query: 7 GVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDLGTTYSCVGV+ +N VEII NDQGNR TPS V+FT+ +RLIG+AAKN AA
Sbjct: 155 GTVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 328
Query: 67 NPSNTIFAFKVIAGT---------------------NDKPTIVVNYK-GEEKRFVAEEIS 104
NP TIF K + G + KP I V K GE K F EE+S
Sbjct: 329 NPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIVNKDGKPYIQVRVKDGETKVFSPEEVS 508
Query: 105 SVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAA 164
++IL+KM+E AEAFL +++AV++VPAYFND+QR+ATKDAG IAGLNV +IINEPTAAA
Sbjct: 509 AMILTKMKETAEAFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAA 688
Query: 165 LAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMV 224
+AYGL K+ ++NI +FDLGGGTFDVSILT+ N VFEV +T GDTHLGG+DFD ++
Sbjct: 689 IAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLSTNGDTHLGGEDFDQRIM 859
Query: 225 DHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSIT 284
++F+K + K+ +DIS++++AL +LR E+AKR LS +E+++++ G+DF +T
Sbjct: 860 EYFIKLIKKKHSKDISKDNRALGKLRRESERAKRALSSQHQVRVEIESLFDGVDFSEPLT 1039
Query: 285 RAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKE 344
RA+FEE+N +LF K M V + DA + KN +D++VLVGGS+RIPKV+QLL++ F GKE
Sbjct: 1040RARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFDGKE 1219
Query: 345 LCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNT 403
K +NPDEAVA+GA VQ ++LS EG + +++L DV PL+ G+ +G +M+ +IPRNT
Sbjct: 1220PNKGVNPDEAVAFGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRNT 1399
Query: 404 SIPVTVEKTKTYYE 417
IP + T Y+
Sbjct: 1400VIPTKKSQVFTTYQ 1441
>TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa protein
(HSP70). {Neurospora crassa}, partial (54%)
Length = 1258
Score = 389 bits (998), Expect(2) = e-111
Identities = 200/334 (59%), Positives = 253/334 (74%), Gaps = 21/334 (6%)
Frame = +2
Query: 9 AIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNP 68
AIGIDLGTTYSCVG + ND++EII NDQGNRTTPS VAF +++RLIGDAAKNQ A NP
Sbjct: 188 AIGIDLGTTYSCVGRY--ANDKIEIIANDQGNRTTPSYVAFNDTERLIGDAAKNQVAMNP 361
Query: 69 SNTIFAFKVIAGTN---------------------DKPTIVVNYKGEEKRFVAEEISSVI 107
NT+F K + G KP I V +KGE K F EEIS+++
Sbjct: 362 HNTVFDAKRLIGRKFSDSEVQADMKHFPFKVIDKGGKPNIEVEFKGENKTFTPEEISAMV 541
Query: 108 LSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALAY 167
L KMRE AEA+L V NAVI+VPAYFNDSQR+ATKDAG IAGLNV++IINEPTAAA+AY
Sbjct: 542 LVKMRETAEAYLGGQVTNAVITVPAYFNDSQRQATKDAGLIAGLNVLRIINEPTAAAIAY 721
Query: 168 GLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMVDHF 227
GL K+A +RN+ IFDLGGGTFDVS+LT++ +FEVK+TAGDTHLGG+DFDN +V+HF
Sbjct: 722 GLDKKAE--GERNVLIFDLGGGTFDVSLLTIEEGIFEVKSTAGDTHLGGEDFDNRLVNHF 895
Query: 228 VKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSITRAK 287
V EF+ K+K+D+S N++ALRRLRTACE+AKRTLS +IE+D++++GIDF +SITRA+
Sbjct: 896 VNEFKRKHKKDLSSNARALRRLRTACERAKRTLSSSAQTSIEIDSLFEGIDFYTSITRAR 1075
Query: 288 FEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVV 321
FEE+ +LF + V+ L+DAK+DK+ V ++V
Sbjct: 1076FEELCQDLFRSTIQPVDRVLSDAKIDKSQVHEIV 1177
Score = 32.7 bits (73), Expect(2) = e-111
Identities = 12/22 (54%), Positives = 19/22 (85%)
Frame = +3
Query: 323 VGGSSRIPKVRQLLQEVFKGKE 344
VGGS+RIP++++L+ + F GKE
Sbjct: 1182 VGGSTRIPRIQKLISDYFNGKE 1247
>TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (44%)
Length = 955
Score = 382 bits (982), Expect = e-106
Identities = 196/284 (69%), Positives = 228/284 (80%), Gaps = 22/284 (7%)
Frame = +2
Query: 6 EGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAA 65
EG AIGIDLGTTYSCVGVW QNDRVEII NDQGNRTTPS VAFT+++RLIGDAAKNQ A
Sbjct: 110 EGKAIGIDLGTTYSCVGVW--QNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKNQVA 283
Query: 66 TNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRFVAEEI 103
NP NT+F FKV+ G +KP IVVNYKGEEK+F AEEI
Sbjct: 284 MNPQNTVFDAKRLIGRRFSDESVQNDMKLWPFKVVPGPAEKPMIVVNYKGEEKKFAAEEI 463
Query: 104 SSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAA 163
SS++L KMRE+AEAFL PVKNAV++VPAYFNDSQR+ATKDAGAI+GLNV++IINEPTAA
Sbjct: 464 SSMVLIKMREVAEAFLGHPVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAA 643
Query: 164 ALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIM 223
A+AYGL K+A+ ++N+ IFDLGGGTFDVS+LT++ +FEVKATAGDTHLGG+DFDN M
Sbjct: 644 AIAYGLDKKASRKGEQNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRM 823
Query: 224 VDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDAT 267
V+HFV EFR K K+DIS N++ALRRLRTACE+AKRTLS T
Sbjct: 824 VNHFVSEFRRKNKKDISGNARALRRLRTACERAKRTLSSTAQTT 955
>TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa protein
mitochondrial precursor. [Kidney bean French bean]
{Phaseolus vulgaris}, complete
Length = 2371
Score = 341 bits (875), Expect = 3e-94
Identities = 205/447 (45%), Positives = 273/447 (60%), Gaps = 23/447 (5%)
Frame = +2
Query: 10 IGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNS-QRLIGDAAKNQAATNP 68
IGIDLGTT SCV V +N +V + N +G RTTPS VAFT + L+G AK QA TNP
Sbjct: 263 IGIDLGTTNSCVSVMEGKNPKV--VENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNP 436
Query: 69 SNTIFAFKVIAGTN------DKPTIVVNYK------------GEEKRFVAEEISSVILSK 110
NTI K + G K +V YK + +++ +I + +L+K
Sbjct: 437 ENTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTK 616
Query: 111 MREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALAYGLQ 170
M+E AEA+L V AVI+VPAYFND+QR+ATKDAG IAGL V++IINEPTAAAL+YG+
Sbjct: 617 MKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMN 796
Query: 171 KRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMVDHFVKE 230
K I +FDLGGGTFDVSIL + N VFEVKAT GDT LGG+DFDN ++D V E
Sbjct: 797 KEGL------IAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNE 958
Query: 231 FRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCS----SITRA 286
F+ D+S++ AL+RLR A EKAK LS + I L I ++TR+
Sbjct: 959 FKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRS 1138
Query: 287 KFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKELC 346
KFE + NL E+ CL DA + +D+V+LVGG +R+PKV++++ +F GK C
Sbjct: 1139KFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-GKSPC 1315
Query: 347 KSINPDEAVAYGAVVQAALLSEGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNTSIP 406
K +NPDEAVA GA +Q +L +V L+L DVTPLS G+ LG + + +I RNT+IP
Sbjct: 1316KGVNPDEAVAMGAALQGGIL---RGDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIP 1486
Query: 407 VTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+K++ + D N+ V +KV Q
Sbjct: 1487--TKKSQVFSTAAD--NQTQVGIKVLQ 1555
>TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chaperone PHSP1
precursor mitochondrial - garden pea, partial (98%)
Length = 2422
Score = 332 bits (851), Expect = 2e-91
Identities = 195/420 (46%), Positives = 262/420 (61%), Gaps = 23/420 (5%)
Frame = +1
Query: 10 IGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNS-QRLIGDAAKNQAATNP 68
IGIDLGTT SCV + +N +V I N +G RTTPS VAF + L+G AK QA TNP
Sbjct: 247 IGIDLGTTNSCVSLMEGKNPKV--IENSEGARTTPSVVAFNQKGELLVGTPAKRQAVTNP 420
Query: 69 SNTIFAFKVIAGTN------DKPTIVVNYK------GE------EKRFVAEEISSVILSK 110
+NT+F K + G K +V YK G+ ++++ +I + +L+K
Sbjct: 421 TNTLFGTKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEINKQQYSPSQIGAFVLTK 600
Query: 111 MREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALAYGLQ 170
M+E AEA+L + AV++VPAYFND+QR+ATKDAG IAGL V +IINEPTAAAL+YG+
Sbjct: 601 MKETAEAYLGKTISKAVVTVPAYFNDAQRQATKDAGRIAGLEVKRIINEPTAAALSYGMN 780
Query: 171 KRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMVDHFVKE 230
+ I +FDLGGGTFDVSIL + N VFEVKAT GDT LGG+DFDN ++D V E
Sbjct: 781 NKEGL-----IAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVSE 945
Query: 231 FRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCS----SITRA 286
F+ D++++ AL+RLR A EKAK LS + I L I ++TR+
Sbjct: 946 FKRTDSIDLAKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRS 1125
Query: 287 KFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKELC 346
KFE + NL E+ CL DA + VD+V+LVGG +R+PKV++++ E+F GK
Sbjct: 1126KFEALVNNLIERTKAPCKSCLKDANISIKDVDEVLLVGGMTRVPKVQEVVSEIF-GKSPS 1302
Query: 347 KSINPDEAVAYGAVVQAALLSEGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNTSIP 406
K +NPDEAVA GA +Q +L +V L+L DVTPLS G+ LG + + +I RNT+IP
Sbjct: 1303KGVNPDEAVAMGAALQGGIL---RGDVKELLLLDVTPLSLGIETLGGIFTRLISRNTTIP 1473
>BQ139103 homologue to GP|2655420|gb| heat shock cognate protein HSC70
{Brassica napus}, partial (29%)
Length = 628
Score = 244 bits (622), Expect = 6e-65
Identities = 130/193 (67%), Positives = 148/193 (76%), Gaps = 22/193 (11%)
Frame = +2
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVW Q+DRVEII NDQGNRTTPS VAFT+S+RLIGDAA
Sbjct: 56 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 229
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
KNQ A NP NT+F FK+I+G +KP I VNYKGE+K F
Sbjct: 230 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKIISGPAEKPLIGVNYKGEDKEF 409
Query: 99 VAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIIN 158
AEEISS++L KMREIAEA+L + +KNAV++VPAYFNDSQR+ATKDAG IAGLNVM+IIN
Sbjct: 410 AAEEISSMVLMKMREIAEAYLGSAIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 589
Query: 159 EPTAAALAYGLQK 171
EPTAAA+AYGL K
Sbjct: 590 EPTAAAIAYGLDK 628
>TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic reticulum
HSC70-cognate binding protein precursor {Glycine max},
partial (38%)
Length = 962
Score = 242 bits (617), Expect = 2e-64
Identities = 135/229 (58%), Positives = 160/229 (68%), Gaps = 22/229 (9%)
Frame = +1
Query: 7 GVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDLGTTYSCVGV+ +N VEII NDQGNR TPS V+FT+ +RLIG+AAKN AA
Sbjct: 289 GTVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 462
Query: 67 NPSNTIFAFKVIAGT---------------------NDKPTIVVNYK-GEEKRFVAEEIS 104
NP TIF K + G + KP I V K GE K F EEIS
Sbjct: 463 NPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNRDGKPYIQVRVKDGETKVFSPEEIS 642
Query: 105 SVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAA 164
++IL KM+E AE FL +++AV++VPAYFND+QR+ATKDAG IAGLNV +IINEPTAAA
Sbjct: 643 AMILGKMKETAEGFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAA 822
Query: 165 LAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTH 213
+AYGL K+ ++NI +FDLGGGTFDVSILT+ N VFEV AT GDTH
Sbjct: 823 IAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTH 960
>TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chaperone
HSC71.0 - garden pea, partial (59%)
Length = 1336
Score = 229 bits (584), Expect = 2e-60
Identities = 119/191 (62%), Positives = 149/191 (77%), Gaps = 1/191 (0%)
Frame = +2
Query: 244 KALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSITRAKFEEMNMNLFEKCMNTV 303
+ALRRLRTACE+AKRTLS TIE+D++++GIDF S ITRA+FEE+NM+LF KCM V
Sbjct: 2 RALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFYSPITRARFEELNMDLFRKCMEPV 181
Query: 304 NICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKELCKSINPDEAVAYGAVVQA 363
CL DAKMDK SV DVVLVGGS+RIPKV+QLLQ+ F GKELCKSINPDEAVAYGA VQA
Sbjct: 182 EKCLRDAKMDKKSVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQA 361
Query: 364 ALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNTSIPVTVEKTKTYYECYDYY 422
A+LS EG++ V +L+L DVTPLS G+ G +M+V+IPRNT+IP E+ + Y
Sbjct: 362 AILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYS----D 529
Query: 423 NKYSVTVKVYQ 433
N+ V ++V++
Sbjct: 530 NQPGVLIQVFE 562
>TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock protein
{Arabidopsis thaliana}, partial (92%)
Length = 3027
Score = 216 bits (551), Expect = 1e-56
Identities = 142/453 (31%), Positives = 225/453 (49%), Gaps = 42/453 (9%)
Frame = +1
Query: 10 IGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNPS 69
+G D G V V ++ ++++ ND+ R TP+ V F + QR IG A NP
Sbjct: 247 VGFDFGNESCIVAVARQRG--IDVVLNDESKRETPAIVCFGDKQRFIGTAGAASTMMNPK 420
Query: 70 NTI----------------------FAFKVIAGTNDKPTIVVNYKGEEKRFVAEEISSVI 107
N+I F V G + P I Y GE + F A ++ ++
Sbjct: 421 NSISQIKRLIGKKFADPELQRDLKSLPFNVTEGPDGYPLIHARYLGESREFTATQVFGMM 600
Query: 108 LSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALAY 167
LS ++EIA+ L V + I +P YF D QRR+ DA IAGL+ + +I+E TA ALAY
Sbjct: 601 LSNLKEIAQKNLNAAVVDCCIGIPVYFTDLQRRSVLDAATIAGLHPLHLIHETTATALAY 780
Query: 168 GLQKRANCVDK-RNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMVDH 226
G+ K ++ N+ D+G + V I K V + + D LGG+DFD + H
Sbjct: 781 GIYKTDLPENEWLNVAFVDVGHASMQVCIAGFKKGQLHVLSHSYDRSLGGRDFDEALFHH 960
Query: 227 FVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSITRA 286
F +F+ +YK D+ +N++A RLR ACEK K+ LS + +A + ++ + D I R
Sbjct: 961 FAAKFKEEYKIDVYQNARACLRLRAACEKLKKVLSANPEAPLNIECLMDEKDVRGFIKRD 1140
Query: 287 KFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKELC 346
FE++++ + E+ + LA+A + ++ V +VG SR+P + ++L E FK KE
Sbjct: 1141DFEQLSLPILERVKGPLEKALAEAGLTVENIHMVEVVGSGSRVPAINKILTEFFK-KEPR 1317
Query: 347 KSINPDEAVAYGAVVQAALLSEGSKNVPNLVLRDVTPLSFGV-----------SELGNLM 395
+++N E VA GA +Q A+LS K V + + P S + SE N
Sbjct: 1318RTMNASECVARGAALQCAILSPTFK-VREFQVNESFPFSVSLSWKYSGSDAPDSESDNKQ 1494
Query: 396 S-VMIPRNTSIP----VTVEKTKTY---YECYD 420
S ++ P+ IP +T +T T+ +C+D
Sbjct: 1495STIVFPKGNPIPSSKVLTFFRTGTFSVDVQCHD 1593
>TC85397 homologue to GP|170386|gb|AAA99920.1|| glucose-regulated protein 78
{Lycopersicon esculentum}, partial (57%)
Length = 648
Score = 211 bits (538), Expect = 3e-55
Identities = 123/246 (50%), Positives = 158/246 (64%)
Frame = +1
Query: 16 TTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNPSNTIFAF 75
TTYSCVGV+ +N VEII NDQGNR TPS V+FT+ +RLIG+AAKN AA NP TIF
Sbjct: 1 TTYSCVGVY--KNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAVNPERTIFDV 174
Query: 76 KVIAGTNDKPTIVVNYKGEEKRFVAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYFN 135
K + G K E+K V ++ V + + ++Q
Sbjct: 175 KRLIGR----------KFEDKE-VQRDMKLVPYKIVNRDGKPYIQ--------------- 276
Query: 136 DSQRRATKDAGAIAGLNVMQIINEPTAAALAYGLQKRANCVDKRNIFIFDLGGGTFDVSI 195
ATKDAG IAGLNV +IINEPTAAA+AYGL K+ ++NI +FDLGGGTFDVSI
Sbjct: 277 -----ATKDAGVIAGLNVARIINEPTAAAIAYGLDKKGG---EKNILVFDLGGGTFDVSI 432
Query: 196 LTVKNNVFEVKATAGDTHLGGQDFDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEK 255
LT+ N VFEV AT GDTHLGG+DFD + ++F+K + K+ +DIS++++AL +LR E+
Sbjct: 433 LTIDNGVFEVLATNGDTHLGGEDFDQRIREYFIKLIKKKHSKDISKDNRALGKLRKESER 612
Query: 256 AKRTLS 261
AKR LS
Sbjct: 613 AKRALS 630
>TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chaperone hsc70-3
- tomato, partial (27%)
Length = 667
Score = 208 bits (530), Expect = 3e-54
Identities = 116/194 (59%), Positives = 138/194 (70%), Gaps = 22/194 (11%)
Frame = +3
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M + + VAIGIDLGTTYSCV V +N ++II ND G RTTPS VAF +S+R+IGDAA
Sbjct: 81 MSGRGKKVAIGIDLGTTYSCVAVC--KNGEIDIIVNDLGKRTTPSFVAFKDSERMIGDAA 254
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
N AA+NP+NTIF FKVI NDKP IVVNY EEK F
Sbjct: 255 FNIAASNPTNTIFDAKRLIGRKFSDPIVQSDVKLWPFKVIGDLNDKPMIVVNYNDEEKHF 434
Query: 99 VAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIIN 158
AEEISS++L KMREIAE FL + V++ VI+VPAYFNDSQR++T+DAGAIAGLNVM+IIN
Sbjct: 435 AAEEISSMVLVKMREIAETFLGSIVEDVVITVPAYFNDSQRQSTRDAGAIAGLNVMRIIN 614
Query: 159 EPTAAALAYGLQKR 172
EPTAAA+AYG +
Sbjct: 615 EPTAAAIAYGFNTK 656
>AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 kDa protein.
[Petunia] {Petunia hybrida}, partial (24%)
Length = 532
Score = 184 bits (467), Expect = 6e-47
Identities = 94/167 (56%), Positives = 118/167 (70%), Gaps = 22/167 (13%)
Frame = +1
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M E A+GIDLGTTYSCV VW E+ +RVEIIHND+G++ TPS VAFT+ QRL+G AA
Sbjct: 31 MANNCECYAVGIDLGTTYSCVAVWLEEKNRVEIIHNDEGSKITPSFVAFTDGQRLVGAAA 210
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
K+QAA NP NT+F FKV++G NDKP I + +KG+EK
Sbjct: 211 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDILLWPFKVVSGVNDKPMISLKFKGQEKLL 390
Query: 99 VAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDA 145
AEEISS++L+ MREIAE +L++PVKNA I+VPAYFND+QR+AT DA
Sbjct: 391 CAEEISSIVLTNMREIAEMYLESPVKNAGITVPAYFNDAQRKATIDA 531
>TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [imported] -
Arabidopsis thaliana, partial (91%)
Length = 1717
Score = 109 bits (273), Expect(2) = 4e-45
Identities = 63/168 (37%), Positives = 96/168 (56%), Gaps = 2/168 (1%)
Frame = +1
Query: 241 ENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSITRAKFEEMNMNLFEKCM 300
E K++ LR A + A LS + +++D + + C + RA+FEE+N +FEKC
Sbjct: 763 EEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKICKVVDRAEFEEVNKEVFEKCE 939
Query: 301 NTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKELCKSINPDEAVAYGAV 360
+ + CL DAK++ ++DV+LVGG S IP+V L+ + K E+ K INP E GA
Sbjct: 940 SLIIQCLHDAKVEVGDINDVILVGGCSYIPRVENLVTNLCKITEVYKGINPLEGAVCGAT 1119
Query: 361 VQAALLSEGSKNVPNLVLRDV--TPLSFGVSELGNLMSVMIPRNTSIP 406
++ A+ S S NL L + TPL+ G+ GN +IPRNT++P
Sbjct: 1120MEGAVASGISDPFGNLDLLTIQATPLAIGIRADGNKFVPVIPRNTTMP 1263
Score = 90.1 bits (222), Expect(2) = 4e-45
Identities = 67/253 (26%), Positives = 115/253 (44%), Gaps = 31/253 (12%)
Frame = +3
Query: 15 GTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFT------------------------ 50
GT+ V VW+ VE++ N + + S V F
Sbjct: 3 GTSQCSVAVWN--GSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFGDTI 176
Query: 51 -NSQRLIGDAAKNQAATNPSNTIFAFKVIAGTNDKPTIVVNYKGEEKRFVAEEISSVILS 109
N +RLIG + N F + + +P I + EE+ ++ L
Sbjct: 177 FNMKRLIGRVDTDPVVHASKNLPFLVQTL-DIGVRPFIAALVNNVWRSTTPEEVLAMFLV 353
Query: 110 KMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALAYGL 169
++R + E L+ P++N V++VP F+ Q + A A+AGL+V++++ EPTA AL YG
Sbjct: 354 ELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALLYGQ 533
Query: 170 QKRANCVD------KRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIM 223
Q++ + ++ IF++G G DV++ V ++KA AG T +GG+D M
Sbjct: 534 QQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDLLQNM 710
Query: 224 VDHFVKEFRNKYK 236
+ H + + N +K
Sbjct: 711 MRHLLPDSENIFK 749
>TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (53%)
Length = 1189
Score = 172 bits (436), Expect = 2e-43
Identities = 90/152 (59%), Positives = 115/152 (75%), Gaps = 1/152 (0%)
Frame = +3
Query: 283 ITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKG 342
ITRA+FEE+NM+LF KCM V CL DAK+DK+ V +VVLVGGS+RIPKV+QLLQ+ F G
Sbjct: 3 ITRARFEELNMDLFRKCMEPVEKCLRDAKIDKSHVHEVVLVGGSTRIPKVQQLLQDFFNG 182
Query: 343 KELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPR 401
KELCKSINPDEAVAYGA VQAA+LS EG + V +L+L DVTPLS G+ G +M+ +IPR
Sbjct: 183 KELCKSINPDEAVAYGAAVQAAILSGEGDEKVQDLLLLDVTPLSLGLETAGGVMTTLIPR 362
Query: 402 NTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
NT+IP E+ + Y N+ V ++V++
Sbjct: 363 NTTIPTKKEQIFSTYS----DNQPGVLIQVFE 446
>TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chaperone BiP-A
- soybean, partial (52%)
Length = 1231
Score = 142 bits (358), Expect = 3e-34
Identities = 70/143 (48%), Positives = 99/143 (68%), Gaps = 1/143 (0%)
Frame = +1
Query: 276 GIDFCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQL 335
G+ F +TRA+FEE+N +LF K M V + DA + KN +D++VLVGGS+RIPKV+QL
Sbjct: 4 GVAFSEPLTRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQL 183
Query: 336 LQEVFKGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNL 394
L++ F GKE K +NPDEAVA+GA VQ ++LS EG +++L DV PL+ G+ +G +
Sbjct: 184 LKDYFDGKEPNKGVNPDEAVAFGAAVQGSILSGEGGAETKDILLLDVAPLTLGIETVGGV 363
Query: 395 MSVMIPRNTSIPVTVEKTKTYYE 417
M+ +IPRNT IP + T Y+
Sbjct: 364 MTKLIPRNTVIPTKKSQVFTTYQ 432
>CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus stolonifer},
partial (19%)
Length = 527
Score = 133 bits (335), Expect = 1e-31
Identities = 74/126 (58%), Positives = 84/126 (65%), Gaps = 21/126 (16%)
Frame = +3
Query: 6 EGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAA 65
EG A+GIDLGTTYSCVGVW QNDRVEII NDQGNRTTPS VAFT+++RLIGDAAKNQ A
Sbjct: 156 EGKAVGIDLGTTYSCVGVW--QNDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVA 329
Query: 66 TNPSNTIFAFKVIAGTN---------------------DKPTIVVNYKGEEKRFVAEEIS 104
NP NT+F K + G KP I V YKGE K+F EEIS
Sbjct: 330 MNPYNTVFDAKRLIGRRFADQEVQSDMKHWPFKVIDKAAKPYIQVEYKGETKQFTPEEIS 509
Query: 105 SVILSK 110
S++L+K
Sbjct: 510 SMVLTK 527
>AW695262 homologue to GP|170386|gb|A glucose-regulated protein 78
{Lycopersicon esculentum}, partial (39%)
Length = 607
Score = 132 bits (333), Expect = 2e-31
Identities = 80/157 (50%), Positives = 100/157 (62%), Gaps = 23/157 (14%)
Frame = +2
Query: 7 GVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDLGTTYSCVGV+ +N VEII NDQGNR TPS V+FT+ +RLIG+AAKN AA
Sbjct: 128 GTVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 301
Query: 67 NPSNTIFAFKVIAG---------------------TNDKPTIVVNYK-GEEKRFVAEEIS 104
NP TIF K + G + KP I V K GE K F EE+S
Sbjct: 302 NPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIVNKDGKPYIQVRVKDGETKVFSPEEVS 481
Query: 105 SVILSKMREIAEAFLQTPVKNAVISVPAYF-NDSQRR 140
++IL+KM+E AEAFL +++AV++VPA ND+QR+
Sbjct: 482 AMILTKMKETAEAFLGKTIRDAVVTVPALLSNDAQRQ 592
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.133 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,533,675
Number of Sequences: 36976
Number of extensions: 119852
Number of successful extensions: 677
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 617
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 619
length of query: 433
length of database: 9,014,727
effective HSP length: 99
effective length of query: 334
effective length of database: 5,354,103
effective search space: 1788270402
effective search space used: 1788270402
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144722.18


