

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.15 + phase: 0
(616 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
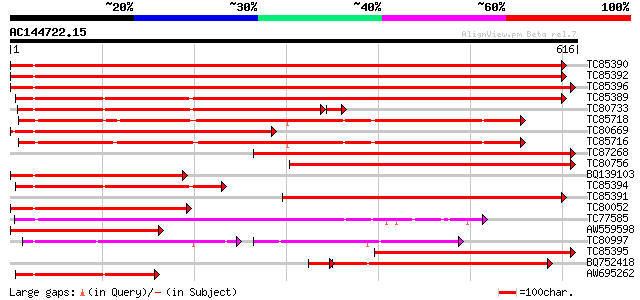
Score E
Sequences producing significant alignments: (bits) Value
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 769 0.0
TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chape... 769 0.0
TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protei... 761 0.0
TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protei... 604 e-173
TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa prot... 425 e-123
TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa prot... 435 e-122
TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 429 e-120
TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chape... 421 e-118
TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chape... 375 e-104
TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 309 2e-84
BQ139103 homologue to GP|2655420|gb| heat shock cognate protein ... 288 3e-78
TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic retic... 281 5e-76
TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chape... 273 1e-73
TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chapero... 257 1e-68
TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock... 249 2e-66
AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 ... 249 3e-66
TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [im... 139 4e-60
TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protei... 206 3e-53
BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated pr... 191 5e-53
AW695262 homologue to GP|170386|gb|A glucose-regulated protein 7... 177 1e-44
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 769 bits (1986), Expect = 0.0
Identities = 395/608 (64%), Positives = 486/608 (78%), Gaps = 3/608 (0%)
Frame = +1
Query: 1 MAKKHKGPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAA 60
MA K +GPAIGIDLGTTYSCV VWQ ++ EII NDQGNR TPS+VAFT+ +RLIGDAA
Sbjct: 136 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 309
Query: 61 KNQAASNPANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRL 120
KNQ A NP NTVFDAKRLIGR+F D +Q+DM+LWPFKV+ G DKP I+V YK EEK+
Sbjct: 310 KNQVAMNPTNTVFDAKRLIGRRFGDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQF 489
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIIN 180
AEEISSMVL KM+EIAE +L + +KNAVVTVPAYFNDSQR+ATKDAGVI+GLNV+RIIN
Sbjct: 490 SAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVLRIIN 669
Query: 181 EPTAAGLAYGLQKRESNIGKRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDTHLGGED 240
EPTAA +AYGL K+ ++ G++NV IFDLGGGTFDVSLLT++ F+VKAT+GDTHLGGED
Sbjct: 670 EPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 849
Query: 241 FDNRMVHHLVKEFRRKNKVDLSGNARALRRLKTACERAKRTLSYDTEATIGIDAIYEGVD 300
FDNRMV+H V+EF+RKNK D+SGN RALRRL+TACERAKRTLS + TI ID+++EG+D
Sbjct: 850 FDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGID 1029
Query: 301 FHLLVTRAKFEQLNRDLFEKCLEIVKNCLANAKMDKNIVDDVVLVGGSSRIPKVQQLLQD 360
F+ +TRA+FE+LN DLF KC+E V+ CL +AKMDKN V DVVLVGGS+RIPKVQQLLQD
Sbjct: 1030FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQD 1209
Query: 361 FFMGKDLYNSINPDEAVAYGAAVQAALLC-DGIKNVPNLVLQDVTPLSLGTSIYDDIMDV 419
FF GK+L SINPDEAVAYGAAVQAA+L +G + V +L+L DVTPLSLG +M V
Sbjct: 1210FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 1389
Query: 420 VIPKNTSIPIRKKQEYVTCYDDQSSVNIDVYEGERMVAIENNLLGLFNLK-VRRAPRGLP 478
+IP+NT+IP +K+Q + T D+Q V I VYEGER +NNLLG F L + APRG+P
Sbjct: 1390LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVP 1569
Query: 479 -IQVCFSIDADGILNVSAEEESSGNKKNITITNENGRLTSEEIERMIQEAEHFKAEDMKH 537
I VCF IDA+GILNVSAE++++G K ITITN+ GRL+ EEIE+M+QEAE +K+ED +H
Sbjct: 1570QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEH 1749
Query: 538 VKKVRAMNALDDYLYDMRKVMKDDSVASKLTSVDKVRINSAMIKGKKLIDDNQPKETYVF 597
KKV A NAL++Y Y+MR +KDD +ASKL++ DK +I A+ + +D NQ E F
Sbjct: 1750KKKVEAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEADEF 1929
Query: 598 VDFLRELK 605
D ++EL+
Sbjct: 1930EDKMKELE 1953
>TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chaperone
hsc70-3 - tomato, complete
Length = 2295
Score = 769 bits (1985), Expect = 0.0
Identities = 394/608 (64%), Positives = 487/608 (79%), Gaps = 3/608 (0%)
Frame = +1
Query: 1 MAKKHKGPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAA 60
MA K +GPAIGIDLGTTYSCV VWQ ++ EII NDQGNR TPS+VAFT+ +RLIGDAA
Sbjct: 130 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 303
Query: 61 KNQAASNPANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRL 120
KNQ A NP NTVFDAKRLIGR+ SD +Q+DM+LWPFKV+ G DKP I+V YK EEK+
Sbjct: 304 KNQVAMNPTNTVFDAKRLIGRRISDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQF 483
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIIN 180
AEEISSMVL KM+EIAE +L + +KNAVVTVPAYFNDSQR+ATKDAGVI+GLNVMRIIN
Sbjct: 484 SAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 663
Query: 181 EPTAAGLAYGLQKRESNIGKRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDTHLGGED 240
EPTAA +AYGL K+ ++ G++NV IFDLGGGTFDVSLLT++ F+VKAT+GDTHLGGED
Sbjct: 664 EPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 843
Query: 241 FDNRMVHHLVKEFRRKNKVDLSGNARALRRLKTACERAKRTLSYDTEATIGIDAIYEGVD 300
FDNRMV+H V+EF+RKNK D+SGN RALRR++TACERAKRTLS + TI ID+++EG+D
Sbjct: 844 FDNRMVNHFVQEFKRKNKKDISGNPRALRRVRTACERAKRTLSSTAQTTIEIDSLFEGID 1023
Query: 301 FHLLVTRAKFEQLNRDLFEKCLEIVKNCLANAKMDKNIVDDVVLVGGSSRIPKVQQLLQD 360
F+ +TRA+FE+LN DLF KC+E V+ CL +AKMDKN VDDVVLVGGS+RIPKVQQLLQD
Sbjct: 1024FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVDDVVLVGGSTRIPKVQQLLQD 1203
Query: 361 FFMGKDLYNSINPDEAVAYGAAVQAALLC-DGIKNVPNLVLQDVTPLSLGTSIYDDIMDV 419
FF GK+L SINPDEAVAYGAAVQAA+L +G + V +L+L DVTPLSLG +M V
Sbjct: 1204FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 1383
Query: 420 VIPKNTSIPIRKKQEYVTCYDDQSSVNIDVYEGERMVAIENNLLGLFNLK-VRRAPRGLP 478
+IP+NT+IP +K+Q + T D+Q V I VYEGER +NNLLG F L + APRG+P
Sbjct: 1384LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVP 1563
Query: 479 -IQVCFSIDADGILNVSAEEESSGNKKNITITNENGRLTSEEIERMIQEAEHFKAEDMKH 537
I VCF IDA+GILNVSAE++++G K ITITN+ GRL+ E+IE+M+QEAE +K+ED +H
Sbjct: 1564QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEH 1743
Query: 538 VKKVRAMNALDDYLYDMRKVMKDDSVASKLTSVDKVRINSAMIKGKKLIDDNQPKETYVF 597
+KV A NAL++Y Y+MR +KDD +ASKL++ DK +I A+ + +D NQ E F
Sbjct: 1744KRKVEAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEADEF 1923
Query: 598 VDFLRELK 605
D ++EL+
Sbjct: 1924EDKMKELE 1947
>TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protein 70
{Cucumis sativus}, complete
Length = 2320
Score = 761 bits (1965), Expect = 0.0
Identities = 393/617 (63%), Positives = 487/617 (78%), Gaps = 3/617 (0%)
Frame = +2
Query: 1 MAKKHKGPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAA 60
MA K +GPAIGIDLGTTYSCV VWQ ++ EII NDQGNR TPS+VAFT+ +RLIGDAA
Sbjct: 107 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 280
Query: 61 KNQAASNPANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRL 120
KNQ A NP NTVFDAKRLIGR+ SD+ +Q+DM+LWPFKV++G +KP I V YKGEEK
Sbjct: 281 KNQVAMNPTNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEEKLF 460
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIIN 180
+EEISSMVL KMREIAE +L +KNAVVTVPAYFNDSQR+ATKDAGVIAGLNV+RIIN
Sbjct: 461 ASEEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 640
Query: 181 EPTAAGLAYGLQKRESNIGKRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDTHLGGED 240
EPTAA +AYGL K+ +++G++NV IFDLGGGTFDVSLLT++ F+VKAT+GDTHLGGED
Sbjct: 641 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 820
Query: 241 FDNRMVHHLVKEFRRKNKVDLSGNARALRRLKTACERAKRTLSYDTEATIGIDAIYEGVD 300
FDNRMV H V+EF+RKNK D+SGN RALRRL+TACERAKRTLS + TI ID+++EGVD
Sbjct: 821 FDNRMVTHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVD 1000
Query: 301 FHLLVTRAKFEQLNRDLFEKCLEIVKNCLANAKMDKNIVDDVVLVGGSSRIPKVQQLLQD 360
F+ +TRA+FE+LN DLF KC+E V+ CL +AKMDK+ V DVVLVGGS+RIPKVQQLLQD
Sbjct: 1001FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQD 1180
Query: 361 FFMGKDLYNSINPDEAVAYGAAVQAALLC-DGIKNVPNLVLQDVTPLSLGTSIYDDIMDV 419
FF GK+L SINPDEAVAYGAAVQAA+L +G + V +L+L DVTPLS G +M V
Sbjct: 1181FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSQGLETAGGVMTV 1360
Query: 420 VIPKNTSIPIRKKQEYVTCYDDQSSVNIDVYEGERMVAIENNLLGLFNLK-VRRAPRGLP 478
+IP+NT+IP +K+Q + T D+Q V I VYEGER +NNLLG F L + APRG+P
Sbjct: 1361LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTKDNNLLGKFELSGIPPAPRGVP 1540
Query: 479 -IQVCFSIDADGILNVSAEEESSGNKKNITITNENGRLTSEEIERMIQEAEHFKAEDMKH 537
I VCF IDA+GILNVSAE++++G K ITITN+ GRL+ EEIE+M+QEAE +K+ED +H
Sbjct: 1541QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEH 1720
Query: 538 VKKVRAMNALDDYLYDMRKVMKDDSVASKLTSVDKVRINSAMIKGKKLIDDNQPKETYVF 597
KKV A N+L++Y Y+MR +KD+ ++SKL+ DK +I A+ + +D NQ E F
Sbjct: 1721KKKVEAKNSLENYAYNMRNTIKDEKISSKLSGGDKKQIEDAIEGAIQWLDANQLAEADEF 1900
Query: 598 VDFLRELKNTFDSALNK 614
D ++EL+ + + K
Sbjct: 1901EDKMKELETICNPIIAK 1951
>TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protein 4
precursor (BiP 4) (78 kDa glucose-regulated protein
homolog 4) (GRP 78-4)., partial (94%)
Length = 2265
Score = 604 bits (1558), Expect = e-173
Identities = 319/604 (52%), Positives = 440/604 (72%), Gaps = 5/604 (0%)
Frame = +2
Query: 7 GPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII NDQGNRITPS+V+FT+D+RLIG+AAKN AA
Sbjct: 155 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 328
Query: 67 NPANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYK-GEEKRLCAEEI 125
NP T+FD KRLIGRKF+D +Q DM+L P+K+V+ D KP I V+ K GE K EE+
Sbjct: 329 NPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIVNK-DGKPYIQVRVKDGETKVFSPEEV 505
Query: 126 SSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIINEPTAA 185
S+M+LTKM+E AE FL +++AVVTVPAYFND+QR+ATKDAGVIAGLNV RIINEPTAA
Sbjct: 506 SAMILTKMKETAEAFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 685
Query: 186 GLAYGLQKRESNIGKRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDTHLGGEDFDNRM 245
+AYGL K+ G++N+ +FDLGGGTFDVS+LT+ F+V +T+GDTHLGGEDFD R+
Sbjct: 686 AIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLSTNGDTHLGGEDFDQRI 856
Query: 246 VHHLVKEFRRKNKVDLSGNARALRRLKTACERAKRTLSYDTEATIGIDAIYEGVDFHLLV 305
+ + +K ++K+ D+S + RAL +L+ ERAKR LS + + I+++++GVDF +
Sbjct: 857 MEYFIKLIKKKHSKDISKDNRALGKLRRESERAKRALSSQHQVRVEIESLFDGVDFSEPL 1036
Query: 306 TRAKFEQLNRDLFEKCLEIVKNCLANAKMDKNIVDDVVLVGGSSRIPKVQQLLQDFFMGK 365
TRA+FE+LN DLF K + VK + +A + KN +D++VLVGGS+RIPKVQQLL+D+F GK
Sbjct: 1037TRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFDGK 1216
Query: 366 DLYNSINPDEAVAYGAAVQAALLC-DGIKNVPNLVLQDVTPLSLGTSIYDDIMDVVIPKN 424
+ +NPDEAVA+GAAVQ ++L +G + +++L DV PL+LG +M +IP+N
Sbjct: 1217EPNKGVNPDEAVAFGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRN 1396
Query: 425 TSIPIRKKQEYVTCYDDQSSVNIDVYEGERMVAIENNLLGLFNLK-VRRAPRGLP-IQVC 482
T IP +K Q + T D Q++V+I V+EGER + + LLG F+L + APRG P I+V
Sbjct: 1397TVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGNFDLSGIPPAPRGTPQIEVT 1576
Query: 483 FSIDADGILNVSAEEESSGNKKNITITNENGRLTSEEIERMIQEAEHFKAEDMKHVKKVR 542
F +DA+GILNV AE++ +G + ITITNE GRL+ EEI+RM++EAE F ED K +++
Sbjct: 1577FEVDANGILNVRAEDKGTGKSEKITITNEKGRLSQEEIDRMVREAEEFAEEDKKVKERID 1756
Query: 543 AMNALDDYLYDMRKVMKD-DSVASKLTSVDKVRINSAMIKGKKLIDDNQPKETYVFVDFL 601
A NAL+ Y+Y+M+ + D D +A KL S +K +I +A+ + + +DDNQ E F + L
Sbjct: 1757ARNALETYVYNMKNQISDKDKLADKLESDEKEKIEAAVKEALEWLDDNQTVEKEEFEEKL 1936
Query: 602 RELK 605
+E++
Sbjct: 1937KEVE 1948
>TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa protein
(HSP70). {Neurospora crassa}, partial (54%)
Length = 1258
Score = 425 bits (1093), Expect(2) = e-123
Identities = 219/335 (65%), Positives = 268/335 (79%)
Frame = +2
Query: 9 AIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAAKNQAASNP 68
AIGIDLGTTYSCV + N+ EII NDQGNR TPS+VAF + +RLIGDAAKNQ A NP
Sbjct: 188 AIGIDLGTTYSCVGRYA--NDKIEIIANDQGNRTTPSYVAFNDTERLIGDAAKNQVAMNP 361
Query: 69 ANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRLCAEEISSM 128
NTVFDAKRLIGRKFSDS +Q DM+ +PFKV+ KP I V++KGE K EEIS+M
Sbjct: 362 HNTVFDAKRLIGRKFSDSEVQADMKHFPFKVIDK-GGKPNIEVEFKGENKTFTPEEISAM 538
Query: 129 VLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIINEPTAAGLA 188
VL KMRE AE +L V NAV+TVPAYFNDSQR+ATKDAG+IAGLNV+RIINEPTAA +A
Sbjct: 539 VLVKMRETAEAYLGGQVTNAVITVPAYFNDSQRQATKDAGLIAGLNVLRIINEPTAAAIA 718
Query: 189 YGLQKRESNIGKRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDTHLGGEDFDNRMVHH 248
YGL K+ G+RNV IFDLGGGTFDVSLLT++ F+VK+T+GDTHLGGEDFDNR+V+H
Sbjct: 719 YGLDKKAE--GERNVLIFDLGGGTFDVSLLTIEEGIFEVKSTAGDTHLGGEDFDNRLVNH 892
Query: 249 LVKEFRRKNKVDLSGNARALRRLKTACERAKRTLSYDTEATIGIDAIYEGVDFHLLVTRA 308
V EF+RK+K DLS NARALRRL+TACERAKRTLS + +I ID+++EG+DF+ +TRA
Sbjct: 893 FVNEFKRKHKKDLSSNARALRRLRTACERAKRTLSSSAQTSIEIDSLFEGIDFYTSITRA 1072
Query: 309 KFEQLNRDLFEKCLEIVKNCLANAKMDKNIVDDVV 343
+FE+L +DLF ++ V L++AK+DK+ V ++V
Sbjct: 1073RFEELCQDLFRSTIQPVDRVLSDAKIDKSQVHEIV 1177
Score = 35.4 bits (80), Expect(2) = e-123
Identities = 13/22 (59%), Positives = 20/22 (90%)
Frame = +3
Query: 345 VGGSSRIPKVQQLLQDFFMGKD 366
VGGS+RIP++Q+L+ D+F GK+
Sbjct: 1182 VGGSTRIPRIQKLISDYFNGKE 1247
>TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa protein
mitochondrial precursor. [Kidney bean French bean]
{Phaseolus vulgaris}, complete
Length = 2371
Score = 435 bits (1119), Expect = e-122
Identities = 250/558 (44%), Positives = 357/558 (63%), Gaps = 7/558 (1%)
Frame = +2
Query: 10 IGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTND-QRLIGDAAKNQAASNP 68
IGIDLGTT SCV+V + +N +++ N +G R TPS VAFT + L+G AK QA +NP
Sbjct: 263 IGIDLGTTNSCVSVMEGKN--PKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNP 436
Query: 69 ANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRLCAEEISSM 128
NT+ AKRLIGR+F D Q +M++ P+K+V + V+ KG++ +I +
Sbjct: 437 ENTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKA--PNGDAWVEAKGQQ--YSPSQIGAF 604
Query: 129 VLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIINEPTAAGLA 188
VLTKM+E AE +L V AV+TVPAYFND+QR+ATKDAG IAGL V+RIINEPTAA L+
Sbjct: 605 VLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALS 784
Query: 189 YGLQKRESNIGKRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDTHLGGEDFDNRMVHH 248
YG+ K + +FDLGGGTFDVS+L + F+VKAT+GDT LGGEDFDN ++
Sbjct: 785 YGMNKEGL------IAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDF 946
Query: 249 LVKEFRRKNKVDLSGNARALRRLKTACERAKRTLSYDTEATIGIDAIYEGVD----FHLL 304
LV EF+R +DLS + AL+RL+ A E+AK LS ++ I + I ++
Sbjct: 947 LVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNIT 1126
Query: 305 VTRAKFEQLNRDLFEKCLEIVKNCLANAKMDKNIVDDVVLVGGSSRIPKVQQLLQDFFMG 364
+TR+KFE L +L E+ ++CL +A + +D+V+LVGG +R+PKVQ+++ F G
Sbjct: 1127LTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-G 1303
Query: 365 KDLYNSINPDEAVAYGAAVQAALLCDGIKNVPNLVLQDVTPLSLGTSIYDDIMDVVIPKN 424
K +NPDEAVA GAA+Q +L +K L+L DVTPLSLG I +I +N
Sbjct: 1304KSPCKGVNPDEAVAMGAALQGGILRGDVK---ELLLLDVTPLSLGIETLGGIFTRLINRN 1474
Query: 425 TSIPIRKKQEYVTCYDDQSSVNIDVYEGERMVAIENNLLGLFNL-KVRRAPRGLP-IQVC 482
T+IP +K Q + T D+Q+ V I V +GER +A +N +LG F L + APRG+P I+V
Sbjct: 1475TTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVT 1654
Query: 483 FSIDADGILNVSAEEESSGNKKNITITNENGRLTSEEIERMIQEAEHFKAEDMKHVKKVR 542
F IDA+GI+ VSA+++S+G ++ ITI + G L+ +EI+ M++EAE +D + +
Sbjct: 1655FDIDANGIVTVSAKDKSTGKEQQITIKSSGG-LSDDEIQNMVKEAELHAQKDQERKSLID 1831
Query: 543 AMNALDDYLYDMRKVMKD 560
N+ D +Y + K + +
Sbjct: 1832IRNSADTSIYSIEKSLSE 1885
>TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (44%)
Length = 955
Score = 429 bits (1104), Expect = e-120
Identities = 218/289 (75%), Positives = 245/289 (84%)
Frame = +2
Query: 1 MAKKHKGPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAA 60
MA K +G AIGIDLGTTYSCV VWQ N+ EII NDQGNR TPS+VAFT+ +RLIGDAA
Sbjct: 98 MATK-EGKAIGIDLGTTYSCVGVWQ--NDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAA 268
Query: 61 KNQAASNPANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRL 120
KNQ A NP NTVFDAKRLIGR+FSD +QNDM+LWPFKVV G +KP I+V YKGEEK+
Sbjct: 269 KNQVAMNPQNTVFDAKRLIGRRFSDESVQNDMKLWPFKVVPGPAEKPMIVVNYKGEEKKF 448
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIIN 180
AEEISSMVL KMRE+AE FL PVKNAVVTVPAYFNDSQR+ATKDAG I+GLNV+RIIN
Sbjct: 449 AAEEISSMVLIKMREVAEAFLGHPVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIIN 628
Query: 181 EPTAAGLAYGLQKRESNIGKRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDTHLGGED 240
EPTAA +AYGL K+ S G++NV IFDLGGGTFDVSLLT++ F+VKAT+GDTHLGGED
Sbjct: 629 EPTAAAIAYGLDKKASRKGEQNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 808
Query: 241 FDNRMVHHLVKEFRRKNKVDLSGNARALRRLKTACERAKRTLSYDTEAT 289
FDNRMV+H V EFRRKNK D+SGNARALRRL+TACERAKRTLS + T
Sbjct: 809 FDNRMVNHFVSEFRRKNKKDISGNARALRRLRTACERAKRTLSSTAQTT 955
>TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chaperone PHSP1
precursor mitochondrial - garden pea, partial (98%)
Length = 2422
Score = 421 bits (1083), Expect = e-118
Identities = 246/560 (43%), Positives = 351/560 (61%), Gaps = 9/560 (1%)
Frame = +1
Query: 10 IGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTND-QRLIGDAAKNQAASNP 68
IGIDLGTT SCV++ + +N ++I N +G R TPS VAF + L+G AK QA +NP
Sbjct: 247 IGIDLGTTNSCVSLMEGKN--PKVIENSEGARTTPSVVAFNQKGELLVGTPAKRQAVTNP 420
Query: 69 ANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRLCAEEISSM 128
NT+F KRLIGR+F D Q +M++ P+K+V + + + +++ +I +
Sbjct: 421 TNTLFGTKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEIN----KQQYSPSQIGAF 588
Query: 129 VLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIINEPTAAGLA 188
VLTKM+E AE +L + AVVTVPAYFND+QR+ATKDAG IAGL V RIINEPTAA L+
Sbjct: 589 VLTKMKETAEAYLGKTISKAVVTVPAYFNDAQRQATKDAGRIAGLEVKRIINEPTAAALS 768
Query: 189 YGLQKRESNIGKRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDTHLGGEDFDNRMVHH 248
YG+ +E I +FDLGGGTFDVS+L + F+VKAT+GDT LGGEDFDN ++
Sbjct: 769 YGMNNKEGLIA-----VFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDF 933
Query: 249 LVKEFRRKNKVDLSGNARALRRLKTACERAKRTLSYDTEATIGIDAIYEGVD----FHLL 304
LV EF+R + +DL+ + AL+RL+ A E+AK LS ++ I + I ++
Sbjct: 934 LVSEFKRTDSIDLAKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNIT 1113
Query: 305 VTRAKFEQLNRDLFEKCLEIVKNCLANAKMDKNIVDDVVLVGGSSRIPKVQQLLQDFFMG 364
+TR+KFE L +L E+ K+CL +A + VD+V+LVGG +R+PKVQ+++ + F G
Sbjct: 1114LTRSKFEALVNNLIERTKAPCKSCLKDANISIKDVDEVLLVGGMTRVPKVQEVVSEIF-G 1290
Query: 365 KDLYNSINPDEAVAYGAAVQAALLCDGIKNVPNLVLQDVTPLSLGTSIYDDIMDVVIPKN 424
K +NPDEAVA GAA+Q +L +K L+L DVTPLSLG I +I +N
Sbjct: 1291KSPSKGVNPDEAVAMGAALQGGILRGDVK---ELLLLDVTPLSLGIETLGGIFTRLISRN 1461
Query: 425 TSIPIRKKQEYVTCYDDQSSVNID--VYEGERMVAIENNLLGLFNL-KVRRAPRGLP-IQ 480
T+IP +K Q + T D+Q+ EG R +A +N LG F+L + APRGLP I+
Sbjct: 1462TTIPTKKSQVFSTAADNQTQRGYXRCSQEGXREMAADNKSLGEFDLVGIPPAPRGLPQIE 1641
Query: 481 VCFSIDADGILNVSAEEESSGNKKNITITNENGRLTSEEIERMIQEAEHFKAEDMKHVKK 540
V F IDA+GI+ VSA+++S+G ++ ITI + G L+ +EI M++EAE D +
Sbjct: 1642VTFDIDANGIVTVSAKDKSTGKEQQITIRSSGG-LSDDEINNMVKEAELHAQRDQERKAL 1818
Query: 541 VRAMNALDDYLYDMRKVMKD 560
+ N+ D +Y + K + +
Sbjct: 1819IDIKNSADTSIYSIEKSLSE 1878
>TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chaperone
HSC71.0 - garden pea, partial (59%)
Length = 1336
Score = 375 bits (963), Expect = e-104
Identities = 199/352 (56%), Positives = 259/352 (73%), Gaps = 3/352 (0%)
Frame = +2
Query: 266 RALRRLKTACERAKRTLSYDTEATIGIDAIYEGVDFHLLVTRAKFEQLNRDLFEKCLEIV 325
RALRRL+TACERAKRTLS + TI ID+++EG+DF+ +TRA+FE+LN DLF KC+E V
Sbjct: 2 RALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFYSPITRARFEELNMDLFRKCMEPV 181
Query: 326 KNCLANAKMDKNIVDDVVLVGGSSRIPKVQQLLQDFFMGKDLYNSINPDEAVAYGAAVQA 385
+ CL +AKMDK V DVVLVGGS+RIPKVQQLLQDFF GK+L SINPDEAVAYGAAVQA
Sbjct: 182 EKCLRDAKMDKKSVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQA 361
Query: 386 ALLC-DGIKNVPNLVLQDVTPLSLGTSIYDDIMDVVIPKNTSIPIRKKQEYVTCYDDQSS 444
A+L +G + V +L+L DVTPLSLG +M V+IP+NT+IP +K+Q + T D+Q
Sbjct: 362 AILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPG 541
Query: 445 VNIDVYEGERMVAIENNLLGLFNLK-VRRAPRGLP-IQVCFSIDADGILNVSAEEESSGN 502
V I V+EGER +NNLLG F L + APRG+P I VCF IDA+GILNVSAE++++G
Sbjct: 542 VLIQVFEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQ 721
Query: 503 KKNITITNENGRLTSEEIERMIQEAEHFKAEDMKHVKKVRAMNALDDYLYDMRKVMKDDS 562
K ITITN+ GRL+ E+IE+M+QEAE +K+ED +H KKV A NAL++Y Y+MR +KD+
Sbjct: 722 KNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTIKDEK 901
Query: 563 VASKLTSVDKVRINSAMIKGKKLIDDNQPKETYVFVDFLRELKNTFDSALNK 614
+A KL S DK +I + + +D NQ E F D ++EL+ + + K
Sbjct: 902 IAGKLDSDDKKKIEDTIEAAIQWLDANQLAEADEFEDKMKELEGVCNPIIAK 1057
>TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (53%)
Length = 1189
Score = 309 bits (792), Expect = 2e-84
Identities = 166/313 (53%), Positives = 225/313 (71%), Gaps = 3/313 (0%)
Frame = +3
Query: 305 VTRAKFEQLNRDLFEKCLEIVKNCLANAKMDKNIVDDVVLVGGSSRIPKVQQLLQDFFMG 364
+TRA+FE+LN DLF KC+E V+ CL +AK+DK+ V +VVLVGGS+RIPKVQQLLQDFF G
Sbjct: 3 ITRARFEELNMDLFRKCMEPVEKCLRDAKIDKSHVHEVVLVGGSTRIPKVQQLLQDFFNG 182
Query: 365 KDLYNSINPDEAVAYGAAVQAALLC-DGIKNVPNLVLQDVTPLSLGTSIYDDIMDVVIPK 423
K+L SINPDEAVAYGAAVQAA+L +G + V +L+L DVTPLSLG +M +IP+
Sbjct: 183 KELCKSINPDEAVAYGAAVQAAILSGEGDEKVQDLLLLDVTPLSLGLETAGGVMTTLIPR 362
Query: 424 NTSIPIRKKQEYVTCYDDQSSVNIDVYEGERMVAIENNLLGLFNLK-VRRAPRGLP-IQV 481
NT+IP +K+Q + T D+Q V I V+EGER +NNLLG F L + APRG+P I V
Sbjct: 363 NTTIPTKKEQIFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELTGIPPAPRGVPQINV 542
Query: 482 CFSIDADGILNVSAEEESSGNKKNITITNENGRLTSEEIERMIQEAEHFKAEDMKHVKKV 541
CF IDA+GILNVSAE++++G K ITITN+ GRL+ EEIE+M+++AE +KAED + KKV
Sbjct: 543 CFDIDANGILNVSAEDKTAGVKNKITITNDKGRLSKEEIEKMVKDAEKYKAEDEEVKKKV 722
Query: 542 RAMNALDDYLYDMRKVMKDDSVASKLTSVDKVRINSAMIKGKKLIDDNQPKETYVFVDFL 601
A N++++Y Y+MR +KD+ + KL+ DK +I A+ + ++ NQ E F D
Sbjct: 723 EAKNSIENYAYNMRNTIKDEKIGGKLSHEDKEKIEKAVEDAIQWLEGNQMAEVDEFEDKQ 902
Query: 602 RELKNTFDSALNK 614
+EL+ + + K
Sbjct: 903 KELEGICNPIIAK 941
>BQ139103 homologue to GP|2655420|gb| heat shock cognate protein HSC70
{Brassica napus}, partial (29%)
Length = 628
Score = 288 bits (738), Expect = 3e-78
Identities = 146/193 (75%), Positives = 165/193 (84%)
Frame = +2
Query: 1 MAKKHKGPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAA 60
MA K +GPAIGIDLGTTYSCV VWQ ++ EII NDQGNR TPS+VAFT+ +RLIGDAA
Sbjct: 56 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 229
Query: 61 KNQAASNPANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRL 120
KNQ A NP NTVFDAKRLIGR+FSD+ +Q+DM+LWPFK++SG +KP I V YKGE+K
Sbjct: 230 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKIISGPAEKPLIGVNYKGEDKEF 409
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIIN 180
AEEISSMVL KMREIAE +L S +KNAVVTVPAYFNDSQR+ATKDAGVIAGLNVMRIIN
Sbjct: 410 AAEEISSMVLMKMREIAEAYLGSAIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 589
Query: 181 EPTAAGLAYGLQK 193
EPTAA +AYGL K
Sbjct: 590 EPTAAAIAYGLDK 628
>TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic reticulum
HSC70-cognate binding protein precursor {Glycine max},
partial (38%)
Length = 962
Score = 281 bits (719), Expect = 5e-76
Identities = 148/230 (64%), Positives = 180/230 (77%), Gaps = 1/230 (0%)
Frame = +1
Query: 7 GPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII NDQGNRITPS+V+FT+D+RLIG+AAKN AA
Sbjct: 289 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 462
Query: 67 NPANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYK-GEEKRLCAEEI 125
NP T+FD KRLIGRKF D +Q DM+L P+K+V+ D KP I V+ K GE K EEI
Sbjct: 463 NPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNR-DGKPYIQVRVKDGETKVFSPEEI 639
Query: 126 SSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIINEPTAA 185
S+M+L KM+E AE FL +++AVVTVPAYFND+QR+ATKDAGVIAGLNV RIINEPTAA
Sbjct: 640 SAMILGKMKETAEGFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 819
Query: 186 GLAYGLQKRESNIGKRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDTH 235
+AYGL K+ G++N+ +FDLGGGTFDVS+LT+ F+V AT+GDTH
Sbjct: 820 AIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTH 960
>TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chaperone BiP-A
- soybean, partial (52%)
Length = 1231
Score = 273 bits (698), Expect = 1e-73
Identities = 147/313 (46%), Positives = 215/313 (67%), Gaps = 4/313 (1%)
Frame = +1
Query: 297 EGVDFHLLVTRAKFEQLNRDLFEKCLEIVKNCLANAKMDKNIVDDVVLVGGSSRIPKVQQ 356
+GV F +TRA+FE+LN DLF K + VK + +A + KN +D++VLVGGS+RIPKVQQ
Sbjct: 1 DGVAFSEPLTRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQ 180
Query: 357 LLQDFFMGKDLYNSINPDEAVAYGAAVQAALLC-DGIKNVPNLVLQDVTPLSLGTSIYDD 415
LL+D+F GK+ +NPDEAVA+GAAVQ ++L +G +++L DV PL+LG
Sbjct: 181 LLKDYFDGKEPNKGVNPDEAVAFGAAVQGSILSGEGGAETKDILLLDVAPLTLGIETVGG 360
Query: 416 IMDVVIPKNTSIPIRKKQEYVTCYDDQSSVNIDVYEGERMVAIENNLLGLFNLK-VRRAP 474
+M +IP+NT IP +K Q + T D Q++V+I V+EGER + + LLG F+L + AP
Sbjct: 361 VMTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAP 540
Query: 475 RGLP-IQVCFSIDADGILNVSAEEESSGNKKNITITNENGRLTSEEIERMIQEAEHFKAE 533
RG P I+V F +DA+GILNV AE++ +G + ITITNE GRL+ EEIERM++EAE F E
Sbjct: 541 RGTPQIEVTFEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIERMVREAEEFAEE 720
Query: 534 DMKHVKKVRAMNALDDYLYDMRKVMKD-DSVASKLTSVDKVRINSAMIKGKKLIDDNQPK 592
D K +++ A NAL+ Y+Y+M+ + D D +A KL S +K +I +A+ + + +DDNQ
Sbjct: 721 DKKVKERIDARNALETYVYNMKNQVSDKDKLADKLESDEKEKIETAVKEALEWLDDNQSV 900
Query: 593 ETYVFVDFLRELK 605
E + + L+E++
Sbjct: 901 EKEEYEEKLKEVE 939
>TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chaperone hsc70-3
- tomato, partial (27%)
Length = 667
Score = 257 bits (656), Expect = 1e-68
Identities = 131/197 (66%), Positives = 155/197 (78%)
Frame = +3
Query: 1 MAKKHKGPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAA 60
M+ + K AIGIDLGTTYSCVAV + N +II ND G R TPSFVAF + +R+IGDAA
Sbjct: 81 MSGRGKKVAIGIDLGTTYSCVAVCK--NGEIDIIVNDLGKRTTPSFVAFKDSERMIGDAA 254
Query: 61 KNQAASNPANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRL 120
N AASNP NT+FDAKRLIGRKFSD ++Q+D++LWPFKV+ +DKP I+V Y EEK
Sbjct: 255 FNIAASNPTNTIFDAKRLIGRKFSDPIVQSDVKLWPFKVIGDLNDKPMIVVNYNDEEKHF 434
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIIN 180
AEEISSMVL KMREIAE FL S V++ V+TVPAYFNDSQR++T+DAG IAGLNVMRIIN
Sbjct: 435 AAEEISSMVLVKMREIAETFLGSIVEDVVITVPAYFNDSQRQSTRDAGAIAGLNVMRIIN 614
Query: 181 EPTAAGLAYGLQKRESN 197
EPTAA +AYG + N
Sbjct: 615 EPTAAAIAYGFNTKPFN 665
>TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock protein
{Arabidopsis thaliana}, partial (92%)
Length = 3027
Score = 249 bits (636), Expect = 2e-66
Identities = 167/532 (31%), Positives = 265/532 (49%), Gaps = 18/532 (3%)
Frame = +1
Query: 6 KGPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAAKNQAA 65
K +G D G VAV + + +++ ND+ R TP+ V F + QR IG A
Sbjct: 235 KMSVVGFDFGNESCIVAVARQRG--IDVVLNDESKRETPAIVCFGDKQRFIGTAGAASTM 408
Query: 66 SNPANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRLCAEEI 125
NP N++ KRLIG+KF+D +Q D++ PF V G D P I +Y GE + A ++
Sbjct: 409 MNPKNSISQIKRLIGKKFADPELQRDLKSLPFNVTEGPDGYPLIHARYLGESREFTATQV 588
Query: 126 SSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIINEPTAA 185
M+L+ ++EIA+K L + V + + +P YF D QR++ DA IAGL+ + +I+E TA
Sbjct: 589 FGMMLSNLKEIAQKNLNAAVVDCCIGIPVYFTDLQRRSVLDAATIAGLHPLHLIHETTAT 768
Query: 186 GLAYGLQKRESNIGK-RNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDTHLGGEDFDNR 244
LAYG+ K + + NV D+G + V + K V + S D LGG DFD
Sbjct: 769 ALAYGIYKTDLPENEWLNVAFVDVGHASMQVCIAGFKKGQLHVLSHSYDRSLGGRDFDEA 948
Query: 245 MVHHLVKEFRRKNKVDLSGNARALRRLKTACERAKRTLSYDTEATIGIDAIYEGVDFHLL 304
+ HH +F+ + K+D+ NARA RL+ ACE+ K+ LS + EA + I+ + + D
Sbjct: 949 LFHHFAAKFKEEYKIDVYQNARACLRLRAACEKLKKVLSANPEAPLNIECLMDEKDVRGF 1128
Query: 305 VTRAKFEQLNRDLFEKCLEIVKNCLANAKMDKNIVDDVVLVGGSSRIPKVQQLLQDFFMG 364
+ R FEQL+ + E+ ++ LA A + + V +VG SR+P + ++L +FF
Sbjct: 1129IKRDDFEQLSLPILERVKGPLEKALAEAGLTVENIHMVEVVGSGSRVPAINKILTEFFK- 1305
Query: 365 KDLYNSINPDEAVAYGAAVQAALLCDGIKNVPNLVLQDVTPLSL-------GTSIYDDIM 417
K+ ++N E VA GAA+Q A+L K V + + P S+ G+ D
Sbjct: 1306KEPRRTMNASECVARGAALQCAILSPTFK-VREFQVNESFPFSVSLSWKYSGSDAPDSES 1482
Query: 418 D-----VVIPKNTSIPIRKKQEYVTCYDDQSSVNIDVYE-GERMVAIENNLLGLFNLKVR 471
D +V PK IP K + SV++ ++ E I +G F K
Sbjct: 1483DNKQSTIVFPKGNPIPSSKVLTFFR--TGTFSVDVQCHDLSETPTKISTYTIGPFQTK-- 1650
Query: 472 RAPRGLPIQVCFSIDADGILNVSA----EEESSGNKKNITITNENGRLTSEE 519
+G ++ ++ GI++V + EEE EN ++ ++E
Sbjct: 1651NGDKG-KVKAKVRLNLHGIVSVESATLFEEEEIEVPVTKEFAEENAKMETDE 1803
Score = 29.6 bits (65), Expect = 3.6
Identities = 24/99 (24%), Positives = 41/99 (41%)
Frame = +3
Query: 518 EEIERMIQEAEHFKAEDMKHVKKVRAMNALDDYLYDMRKVMKDDSVASKLTSVDKVRINS 577
E +E +Q+ + +D K NA++ Y+YDMR + D + S I
Sbjct: 2052 EGVEMALQDRVMEETKDNK--------NAIEAYVYDMRNKLNDKYQEFVVASERDGFITK 2207
Query: 578 AMIKGKKLIDDNQPKETYVFVDFLRELKNTFDSALNKFK 616
L +D + + V++ L ELK D ++K
Sbjct: 2208 LQEVEDWLYEDGEDETKGVYIAKLEELKKQGDPIEERYK 2324
>AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 kDa protein.
[Petunia] {Petunia hybrida}, partial (24%)
Length = 532
Score = 249 bits (635), Expect = 3e-66
Identities = 123/167 (73%), Positives = 143/167 (84%)
Frame = +1
Query: 1 MAKKHKGPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAA 60
MA + A+GIDLGTTYSCVAVW ++ N EIIHND+G++ITPSFVAFT+ QRL+G AA
Sbjct: 31 MANNCECYAVGIDLGTTYSCVAVWLEEKNRVEIIHNDEGSKITPSFVAFTDGQRLVGAAA 210
Query: 61 KNQAASNPANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRL 120
K+QAA NP NTVFDAKRLIGRKFSD V+Q D+ LWPFKVVSG +DKP I +K+KG+EK L
Sbjct: 211 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDILLWPFKVVSGVNDKPMISLKFKGQEKLL 390
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDA 167
CAEEISS+VLT MREIAE +LESPVKNA +TVPAYFND+QRKAT DA
Sbjct: 391 CAEEISSIVLTNMREIAEMYLESPVKNAGITVPAYFNDAQRKATIDA 531
>TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [imported] -
Arabidopsis thaliana, partial (91%)
Length = 1717
Score = 139 bits (349), Expect(2) = 4e-60
Identities = 83/233 (35%), Positives = 136/233 (57%), Gaps = 5/233 (2%)
Frame = +1
Query: 266 RALRRLKTACERAKRTLSYDTEATIGIDAIYEGVDFHLLVTRAKFEQLNRDLFEKCLEIV 325
+++ L+ A + A LS + + +D + + +V RA+FE++N+++FEKC ++
Sbjct: 772 KSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKICKVVDRAEFEEVNKEVFEKCESLI 948
Query: 326 KNCLANAKMDKNIVDDVVLVGGSSRIPKVQQLLQDFFMGKDLYNSINPDEAVAYGAAVQA 385
CL +AK++ ++DV+LVGG S IP+V+ L+ + ++Y INP E GA ++
Sbjct: 949 IQCLHDAKVEVGDINDVILVGGCSYIPRVENLVTNLCKITEVYKGINPLEGAVCGATMEG 1128
Query: 386 AL---LCDGIKNVPNLVLQDVTPLSLGTSIYDDIMDVVIPKNTSIPIRKKQEYVTCYDDQ 442
A+ + D N+ L +Q TPL++G + VIP+NT++P RK + T +D+Q
Sbjct: 1129 AVASGISDPFGNLDLLTIQ-ATPLAIGIRADGNKFVPVIPRNTTMPARKDLLFTTIHDNQ 1305
Query: 443 SSVNIDVYEGERMVAIENNLLGLFNLK-VRRAPRGLP-IQVCFSIDADGILNV 493
+ I VYEGE A EN+LLG F + + AP+G+P I VC IDA +L V
Sbjct: 1306 TEALILVYEGEGKKAEENHLLGYFKIMGIPAAPKGVPEISVCMDIDAANVLRV 1464
Score = 111 bits (278), Expect(2) = 4e-60
Identities = 76/247 (30%), Positives = 130/247 (51%), Gaps = 9/247 (3%)
Frame = +3
Query: 15 GTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAAK--NQAASNPANTV 72
GT+ VAVW + E++ N + ++ SFV F ++ G ++ ++ +T+
Sbjct: 3 GTSQCSVAVWN--GSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFGDTI 176
Query: 73 FDAKRLIGRKFSDSVIQNDMELWPFKVVS-GFDDKPEIIVKYKGEEKRLCAEEISSMVLT 131
F+ KRLIGR +D V+ L PF V + +P I + EE+ +M L
Sbjct: 177 FNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAMFLV 353
Query: 132 KMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIINEPTAAGLAYGL 191
++R + E L+ P++N V+TVP F+ Q + A +AGL+V+R++ EPTA L YG
Sbjct: 354 ELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALLYGQ 533
Query: 192 QKRESNI------GKRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDTHLGGEDFDNRM 245
Q+++++ ++ IF++G G DV++ G +KA +G T +GGED M
Sbjct: 534 QQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDLLQNM 710
Query: 246 VHHLVKE 252
+ HL+ +
Sbjct: 711 MRHLLPD 731
>TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protein 70
{Cucumis sativus}, partial (35%)
Length = 723
Score = 206 bits (523), Expect = 3e-53
Identities = 110/220 (50%), Positives = 153/220 (69%), Gaps = 2/220 (0%)
Frame = +3
Query: 397 NLVLQDVTPLSLGTSIYDDIMDVVIPKNTSIPIRKKQEYVTCYDDQSSVNIDVYEGERMV 456
+L+L DVTPLSLG +M V+IP+NT+IP +K+Q + T D+Q V I VYEGER
Sbjct: 6 DLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERAR 185
Query: 457 AIENNLLGLFNLK-VRRAPRGLP-IQVCFSIDADGILNVSAEEESSGNKKNITITNENGR 514
+NNLLG F L + APRG+P I VCF IDA+GILNVSAE++++G K ITITN+ GR
Sbjct: 186 TRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGR 365
Query: 515 LTSEEIERMIQEAEHFKAEDMKHVKKVRAMNALDDYLYDMRKVMKDDSVASKLTSVDKVR 574
L+ EEIE+M++EAE +KAED +H KKV A NAL++Y Y+MR +KD+ + SKL+ DK +
Sbjct: 366 LSKEEIEKMVEEAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIGSKLSPEDKKK 545
Query: 575 INSAMIKGKKLIDDNQPKETYVFVDFLRELKNTFDSALNK 614
I+ A+ + +D NQ E F D ++EL++ + + K
Sbjct: 546 IDDAIDAAIQWLDSNQLAEADEFQDKMKELESLCNPIIAK 665
>BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated protein
homolog precursor (GRP 78), partial (47%)
Length = 1032
Score = 191 bits (486), Expect(2) = 5e-53
Identities = 105/242 (43%), Positives = 155/242 (63%), Gaps = 3/242 (1%)
Frame = -1
Query: 351 IPKVQQLLQDFFMGKDLYNSINPDEAVAYGAAVQAALLCDGIKNVPNLVLQDVTPLSLGT 410
IPKVQ L++++F GK INPDEAVA+GAAVQA +L G + +VL DV PL+LG
Sbjct: 900 IPKVQSLIEEYFGGKKASKGINPDEAVAFGAAVQAGVL-SGEEGTEEIVLMDVNPLTLGI 724
Query: 411 SIYDDIMDVVIPKNTSIPIRKKQEYVTCYDDQSSVNIDVYEGERMVAIENNLLGLFNLK- 469
+M +I +NT IP RK Q + T D+Q V I V+EGER + +NN LG F L
Sbjct: 723 ETTGGVMTKLITRNTPIPTRKSQIFSTAADNQPVVLIQVFEGERSLTKDNNQLGKFELTG 544
Query: 470 VRRAPRGLP-IQVCFSIDADGILNVSAEEESSGNKKNITITNENGRLTSEEIERMIQEAE 528
+ APRG+P I+V F +DA+GIL VSA ++ +G +++ITITN+ GRLT EEI+RM++EAE
Sbjct: 543 IPPAPRGVPQIEVSFELDANGILKVSAHDKGTGKQESITITNDKGRLTKEEIDRMVEEAE 364
Query: 529 HFKAEDMKHVKKVRAMNALDDYLYDMRKVMKD-DSVASKLTSVDKVRINSAMIKGKKLID 587
+ ED +++ A N L++Y + ++ + D + + K+ DK I A+ + ++
Sbjct: 363 KYAEEDKATRERIEARNGLENYAFSLKNQVNDEEGLGGKIDDEDKETILEAVKETNDWLE 184
Query: 588 DN 589
+N
Sbjct: 183 EN 178
Score = 35.0 bits (79), Expect(2) = 5e-53
Identities = 16/29 (55%), Positives = 22/29 (75%)
Frame = -3
Query: 325 VKNCLANAKMDKNIVDDVVLVGGSSRIPK 353
V+ L +AK+ K VDD+VLVGGS+R P+
Sbjct: 979 VEQVLKDAKVKKEDVDDIVLVGGSTRYPQ 893
>AW695262 homologue to GP|170386|gb|A glucose-regulated protein 78
{Lycopersicon esculentum}, partial (39%)
Length = 607
Score = 177 bits (448), Expect = 1e-44
Identities = 96/158 (60%), Positives = 120/158 (75%), Gaps = 2/158 (1%)
Frame = +2
Query: 7 GPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII NDQGNRITPS+V+FT+D+RLIG+AAKN AA
Sbjct: 128 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 301
Query: 67 NPANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYK-GEEKRLCAEEI 125
NP T+FD KRLIGRKF+D +Q DM+L P+K+V+ D KP I V+ K GE K EE+
Sbjct: 302 NPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIVNK-DGKPYIQVRVKDGETKVFSPEEV 478
Query: 126 SSMVLTKMREIAEKFLESPVKNAVVTVPAYF-NDSQRK 162
S+M+LTKM+E AE FL +++AVVTVPA ND+QR+
Sbjct: 479 SAMILTKMKETAEAFLGKTIRDAVVTVPALLSNDAQRQ 592
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.135 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,714,870
Number of Sequences: 36976
Number of extensions: 167359
Number of successful extensions: 810
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 736
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 739
length of query: 616
length of database: 9,014,727
effective HSP length: 102
effective length of query: 514
effective length of database: 5,243,175
effective search space: 2694991950
effective search space used: 2694991950
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144722.15


