

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.13 + phase: 0
(600 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
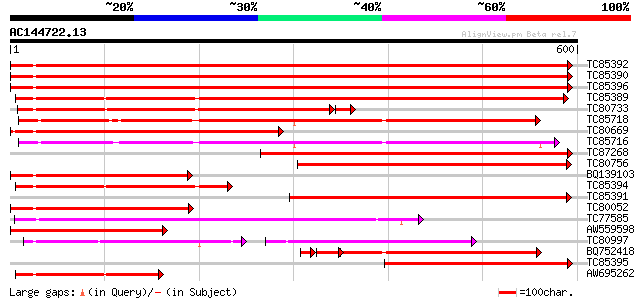
Score E
Sequences producing significant alignments: (bits) Value
TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chape... 780 0.0
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 779 0.0
TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protei... 771 0.0
TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protei... 610 e-175
TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa prot... 441 e-127
TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa prot... 442 e-124
TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 434 e-122
TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chape... 419 e-117
TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chape... 376 e-104
TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 311 5e-85
BQ139103 homologue to GP|2655420|gb| heat shock cognate protein ... 291 4e-79
TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic retic... 288 4e-78
TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chape... 269 2e-72
TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chapero... 264 8e-71
TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock... 247 1e-65
AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 ... 243 1e-64
TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [im... 139 2e-60
BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated pr... 192 1e-55
TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protei... 197 1e-50
AW695262 homologue to GP|170386|gb|A glucose-regulated protein 7... 177 8e-45
>TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chaperone
hsc70-3 - tomato, complete
Length = 2295
Score = 780 bits (2013), Expect = 0.0
Identities = 395/597 (66%), Positives = 480/597 (80%), Gaps = 2/597 (0%)
Frame = +1
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
MA K +GPAIGIDLGTTYSCV VWQ ++R EII NDQGNRTTPS+VAFTD +RLIGDAA
Sbjct: 130 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 303
Query: 61 KNQAASNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHF 120
KNQ A NP NT+FDAKRLIGR+ SD +Q+D++LWPFKV+PG DKP I+V YK EK F
Sbjct: 304 KNQVAMNPTNTVFDAKRLIGRRISDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQF 483
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIIN 180
AEEISSMVL KM+EIAE +L + +KNAVVTVPAYFNDSQR+ATKDAG I+GLNVMRIIN
Sbjct: 484 SAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 663
Query: 181 EPTAAALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGED 240
EPTAAA+AYGL K+A E+N+ IFDLGGGTFDVSLLTI+ +FEVKAT+GDTHLGGED
Sbjct: 664 EPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 843
Query: 241 FDNRMVNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID 300
FDNRMVNHFV EF RK KDISG+PR LRR+RTACER KRTLS TI+IDSLFEGID
Sbjct: 844 FDNRMVNHFVQEFKRKNKKDISGNPRALRRVRTACERAKRTLSSTAQTTIEIDSLFEGID 1023
Query: 301 FHSSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKE 360
F+++ITRA+FE+LN+DLF+KC+E VE CL DA+++K+SVDD VLVGGS+RIPKVQQLL++
Sbjct: 1024FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVDDVVLVGGSTRIPKVQQLLQD 1203
Query: 361 FFNWKDICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSV 419
FFN K++C SINPDEAVAYGAAVQAA+L E EK +L+L DVTPLSLG G +++V
Sbjct: 1204FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 1383
Query: 420 VIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQI 478
+IPRNT IP KK + + T D+ V ++VYEGER + +NNLLGKF S IPPAPRG
Sbjct: 1384LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVP 1563
Query: 479 PIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKF 538
I VCF ID +GILNVSA + T G + +ITITN+KGRLS E+IE+M+QEAE +K ED +
Sbjct: 1564QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEH 1743
Query: 539 KKKVKAMNDLDDYLYTMRKVMKDDSVSSMLTSIERMKINSAMIKGMKLIDEKQHHEA 595
K+KV+A N L++Y Y MR +KDD ++S L++ ++ KI A+ ++ +D Q EA
Sbjct: 1744KRKVEAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEA 1914
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 779 bits (2012), Expect = 0.0
Identities = 395/597 (66%), Positives = 479/597 (80%), Gaps = 2/597 (0%)
Frame = +1
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
MA K +GPAIGIDLGTTYSCV VWQ ++R EII NDQGNRTTPS+VAFTD +RLIGDAA
Sbjct: 136 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 309
Query: 61 KNQAASNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHF 120
KNQ A NP NT+FDAKRLIGR++ D +Q+D++LWPFKV+PG DKP I+V YK EK F
Sbjct: 310 KNQVAMNPTNTVFDAKRLIGRRFGDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQF 489
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIIN 180
AEEISSMVL KM+EIAE +L + +KNAVVTVPAYFNDSQR+ATKDAG I+GLNV+RIIN
Sbjct: 490 SAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVLRIIN 669
Query: 181 EPTAAALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGED 240
EPTAAA+AYGL K+A E+N+ IFDLGGGTFDVSLLTI+ +FEVKAT+GDTHLGGED
Sbjct: 670 EPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 849
Query: 241 FDNRMVNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID 300
FDNRMVNHFV EF RK KDISG+PR LRRLRTACER KRTLS TI+IDSLFEGID
Sbjct: 850 FDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGID 1029
Query: 301 FHSSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKE 360
F+++ITRA+FE+LN+DLF+KC+E VE CL DA+++K+SV D VLVGGS+RIPKVQQLL++
Sbjct: 1030FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQD 1209
Query: 361 FFNWKDICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSV 419
FFN K++C SINPDEAVAYGAAVQAA+L E EK +L+L DVTPLSLG G +++V
Sbjct: 1210FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 1389
Query: 420 VIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQI 478
+IPRNT IP KK + + T D+ V ++VYEGER + +NNLLGKF S IPPAPRG
Sbjct: 1390LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVP 1569
Query: 479 PIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKF 538
I VCF ID +GILNVSA + T G + +ITITN+KGRLS EEIE+M+QEAE +K ED +
Sbjct: 1570QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEH 1749
Query: 539 KKKVKAMNDLDDYLYTMRKVMKDDSVSSMLTSIERMKINSAMIKGMKLIDEKQHHEA 595
KKKV+A N L++Y Y MR +KDD ++S L++ ++ KI A+ ++ +D Q EA
Sbjct: 1750KKKVEAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEA 1920
>TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protein 70
{Cucumis sativus}, complete
Length = 2320
Score = 771 bits (1990), Expect = 0.0
Identities = 393/597 (65%), Positives = 476/597 (78%), Gaps = 2/597 (0%)
Frame = +2
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
MA K +GPAIGIDLGTTYSCV VWQ ++R EII NDQGNRTTPS+VAFTD +RLIGDAA
Sbjct: 107 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 280
Query: 61 KNQAASNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHF 120
KNQ A NP NT+FDAKRLIGR+ SD+ +Q+D++LWPFKV+ G +KP I V YKG EK F
Sbjct: 281 KNQVAMNPTNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEEKLF 460
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIIN 180
+EEISSMVL KMREIAE +L +KNAVVTVPAYFNDSQR+ATKDAG IAGLNV+RIIN
Sbjct: 461 ASEEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 640
Query: 181 EPTAAALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGED 240
EPTAAA+AYGL K+A V E+N+ IFDLGGGTFDVSLLTI+ +FEVKAT+GDTHLGGED
Sbjct: 641 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 820
Query: 241 FDNRMVNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID 300
FDNRMV HFV EF RK KDISG+PR LRRLRTACER KRTLS TI+IDSLFEG+D
Sbjct: 821 FDNRMVTHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVD 1000
Query: 301 FHSSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKE 360
F+++ITRA+FE+LN+DLF+KC+E VE CL DA+++KS+V D VLVGGS+RIPKVQQLL++
Sbjct: 1001FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQD 1180
Query: 361 FFNWKDICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSV 419
FFN K++C SINPDEAVAYGAAVQAA+L E EK +L+L DVTPLS G G +++V
Sbjct: 1181FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSQGLETAGGVMTV 1360
Query: 420 VIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQI 478
+IPRNT IP KK + + T D+ V ++VYEGER + +NNLLGKF S IPPAPRG
Sbjct: 1361LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTKDNNLLGKFELSGIPPAPRGVP 1540
Query: 479 PIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKF 538
I VCF ID +GILNVSA + T G + +ITITN+KGRLS EEIE+M+QEAE +K ED +
Sbjct: 1541QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEH 1720
Query: 539 KKKVKAMNDLDDYLYTMRKVMKDDSVSSMLTSIERMKINSAMIKGMKLIDEKQHHEA 595
KKKV+A N L++Y Y MR +KD+ +SS L+ ++ +I A+ ++ +D Q EA
Sbjct: 1721KKKVEAKNSLENYAYNMRNTIKDEKISSKLSGGDKKQIEDAIEGAIQWLDANQLAEA 1891
>TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protein 4
precursor (BiP 4) (78 kDa glucose-regulated protein
homolog 4) (GRP 78-4)., partial (94%)
Length = 2265
Score = 610 bits (1573), Expect = e-175
Identities = 320/589 (54%), Positives = 430/589 (72%), Gaps = 4/589 (0%)
Frame = +2
Query: 7 GPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII NDQGNR TPS+V+FTDD+RLIG+AAKN AA
Sbjct: 155 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 328
Query: 67 NPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGE-KHFCAEEI 125
NP TIFD KRLIGRK++D +Q D++L P+K+V D KP I V+ K GE K F EE+
Sbjct: 329 NPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIV-NKDGKPYIQVRVKDGETKVFSPEEV 505
Query: 126 SSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAA 185
S+M+LTKM+E AE FL +++AVVTVPAYFND+QR+ATKDAG IAGLNV RIINEPTAA
Sbjct: 506 SAMILTKMKETAEAFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 685
Query: 186 ALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRM 245
A+AYGL K+ E+NI +FDLGGGTFDVS+LTI N +FEV +T+GDTHLGGEDFD R+
Sbjct: 686 AIAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLSTNGDTHLGGEDFDQRI 856
Query: 246 VNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGIDFHSSI 305
+ +F+ +K+ KDIS R L +LR ER KR LS ++I+SLF+G+DF +
Sbjct: 857 MEYFIKLIKKKHSKDISKDNRALGKLRRESERAKRALSSQHQVRVEIESLFDGVDFSEPL 1036
Query: 306 TRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNWK 365
TRA+FE+LN DLF+K + V+ + DA ++K+ +D+ VLVGGS+RIPKVQQLLK++F+ K
Sbjct: 1037TRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFDGK 1216
Query: 366 DICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSVVIPRN 424
+ +NPDEAVA+GAAVQ ++L E E++ +++L DV PL+LG G +++ +IPRN
Sbjct: 1217EPNKGVNPDEAVAFGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRN 1396
Query: 425 TPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQIPIKVC 483
T IP KK++ + T +D + VS++V+EGER + LLG F S IPPAPRG I+V
Sbjct: 1397TVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGNFDLSGIPPAPRGTPQIEVT 1576
Query: 484 FSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKFKKKVK 543
F +D +GILNV A + G ++ITITNEKGRLS EEI+RM++EAE F +ED K K+++
Sbjct: 1577FEVDANGILNVRAEDKGTGKSEKITITNEKGRLSQEEIDRMVREAEEFAEEDKKVKERID 1756
Query: 544 AMNDLDDYLYTMRKVMKD-DSVSSMLTSIERMKINSAMIKGMKLIDEKQ 591
A N L+ Y+Y M+ + D D ++ L S E+ KI +A+ + ++ +D+ Q
Sbjct: 1757ARNALETYVYNMKNQISDKDKLADKLESDEKEKIEAAVKEALEWLDDNQ 1903
>TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa protein
(HSP70). {Neurospora crassa}, partial (54%)
Length = 1258
Score = 441 bits (1133), Expect(2) = e-127
Identities = 228/335 (68%), Positives = 270/335 (80%)
Frame = +2
Query: 9 AIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAASNP 68
AIGIDLGTTYSCV + N++ EII NDQGNRTTPS+VAF D +RLIGDAAKNQ A NP
Sbjct: 188 AIGIDLGTTYSCVGRYA--NDKIEIIANDQGNRTTPSYVAFNDTERLIGDAAKNQVAMNP 361
Query: 69 VNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHFCAEEISSM 128
NT+FDAKRLIGRK+SDS +Q D++ +PFKV+ KP I V++KG K F EEIS+M
Sbjct: 362 HNTVFDAKRLIGRKFSDSEVQADMKHFPFKVID-KGGKPNIEVEFKGENKTFTPEEISAM 538
Query: 129 VLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAALA 188
VL KMRE AE +L V NAV+TVPAYFNDSQR+ATKDAG IAGLNV+RIINEPTAAA+A
Sbjct: 539 VLVKMRETAEAYLGGQVTNAVITVPAYFNDSQRQATKDAGLIAGLNVLRIINEPTAAAIA 718
Query: 189 YGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMVNH 248
YGL K+A ERN+ IFDLGGGTFDVSLLTI+ +FEVK+T+GDTHLGGEDFDNR+VNH
Sbjct: 719 YGLDKKAE--GERNVLIFDLGGGTFDVSLLTIEEGIFEVKSTAGDTHLGGEDFDNRLVNH 892
Query: 249 FVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGIDFHSSITRA 308
FV EF RK+ KD+S + R LRRLRTACER KRTLS +I+IDSLFEGIDF++SITRA
Sbjct: 893 FVNEFKRKHKKDLSSNARALRRLRTACERAKRTLSSSAQTSIEIDSLFEGIDFYTSITRA 1072
Query: 309 KFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAV 343
+FE+L DLF+ I+ V+ LSDA+I+KS V + V
Sbjct: 1073RFEELCQDLFRSTIQPVDRVLSDAKIDKSQVHEIV 1177
Score = 33.9 bits (76), Expect(2) = e-127
Identities = 12/22 (54%), Positives = 20/22 (90%)
Frame = +3
Query: 345 VGGSSRIPKVQQLLKEFFNWKD 366
VGGS+RIP++Q+L+ ++FN K+
Sbjct: 1182 VGGSTRIPRIQKLISDYFNGKE 1247
>TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa protein
mitochondrial precursor. [Kidney bean French bean]
{Phaseolus vulgaris}, complete
Length = 2371
Score = 442 bits (1136), Expect = e-124
Identities = 253/558 (45%), Positives = 355/558 (63%), Gaps = 6/558 (1%)
Frame = +2
Query: 10 IGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDD-KRLIGDAAKNQAASNP 68
IGIDLGTT SCV+V + +N + ++ N +G RTTPS VAFT + L+G AK QA +NP
Sbjct: 263 IGIDLGTTNSCVSVMEGKNPK--VVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNP 436
Query: 69 VNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHFCAEEISSM 128
NTI AKRLIGR++ D Q ++++ P+K+V + + V+ KG + + +I +
Sbjct: 437 ENTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNG--DAWVEAKG--QQYSPSQIGAF 604
Query: 129 VLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAALA 188
VLTKM+E AE +L V AV+TVPAYFND+QR+ATKDAG IAGL V+RIINEPTAAAL+
Sbjct: 605 VLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALS 784
Query: 189 YGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMVNH 248
YG+ K E I +FDLGGGTFDVS+L I N +FEVKAT+GDT LGGEDFDN +++
Sbjct: 785 YGMNK------EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDF 946
Query: 249 FVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID----FHSS 304
V EF R D+S L+RLR A E+ K LS + I++ + + +
Sbjct: 947 LVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNIT 1126
Query: 305 ITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNW 364
+TR+KFE L +L ++ +SCL DA I +D+ +LVGG +R+PKVQ+++ F
Sbjct: 1127LTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFG- 1303
Query: 365 KDICVSINPDEAVAYGAAVQAALLCEATEKSLNLVLRDVTPLSLGTLVRGDLLSVVIPRN 424
K C +NPDEAVA GAA+Q +L ++ L+L DVTPLSLG G + + +I RN
Sbjct: 1304KSPCKGVNPDEAVAMGAALQGGILRGDVKE---LLLLDVTPLSLGIETLGGIFTRLINRN 1474
Query: 425 TPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRF-SIPPAPRGQIPIKVC 483
T IP KK++ + T D+ + V ++V +GER AS+N +LG+F +PPAPRG I+V
Sbjct: 1475TTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVT 1654
Query: 484 FSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKFKKKVK 543
F ID +GI+ VSA + + G Q+ITI G LS +EI+ M++EAE +D + K +
Sbjct: 1655FDIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLID 1831
Query: 544 AMNDLDDYLYTMRKVMKD 561
N D +Y++ K + +
Sbjct: 1832IRNSADTSIYSIEKSLSE 1885
>TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (44%)
Length = 955
Score = 434 bits (1116), Expect = e-122
Identities = 220/289 (76%), Positives = 245/289 (84%)
Frame = +2
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
MA K +G AIGIDLGTTYSCV VWQ N+R EII NDQGNRTTPS+VAFTD +RLIGDAA
Sbjct: 98 MATK-EGKAIGIDLGTTYSCVGVWQ--NDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAA 268
Query: 61 KNQAASNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHF 120
KNQ A NP NT+FDAKRLIGR++SD +QND++LWPFKVVPG +KP I+V YKG EK F
Sbjct: 269 KNQVAMNPQNTVFDAKRLIGRRFSDESVQNDMKLWPFKVVPGPAEKPMIVVNYKGEEKKF 448
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIIN 180
AEEISSMVL KMRE+AE FL PVKNAVVTVPAYFNDSQR+ATKDAGAI+GLNV+RIIN
Sbjct: 449 AAEEISSMVLIKMREVAEAFLGHPVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIIN 628
Query: 181 EPTAAALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGED 240
EPTAAA+AYGL K+A+ E+N+ IFDLGGGTFDVSLLTI+ +FEVKAT+GDTHLGGED
Sbjct: 629 EPTAAAIAYGLDKKASRKGEQNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 808
Query: 241 FDNRMVNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNAT 289
FDNRMVNHFV EF RK KDISG+ R LRRLRTACER KRTLS T
Sbjct: 809 FDNRMVNHFVSEFRRKNKKDISGNARALRRLRTACERAKRTLSSTAQTT 955
>TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chaperone PHSP1
precursor mitochondrial - garden pea, partial (98%)
Length = 2422
Score = 419 bits (1076), Expect = e-117
Identities = 255/597 (42%), Positives = 356/597 (58%), Gaps = 24/597 (4%)
Frame = +1
Query: 10 IGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDD-KRLIGDAAKNQAASNP 68
IGIDLGTT SCV++ + +N + +I N +G RTTPS VAF + L+G AK QA +NP
Sbjct: 247 IGIDLGTTNSCVSLMEGKNPK--VIENSEGARTTPSVVAFNQKGELLVGTPAKRQAVTNP 420
Query: 69 VNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVV--PGNDDKPEIIVKYKGGEKHFCAEEIS 126
NT+F KRLIGR++ D Q ++++ P+K+V P D EI ++ + +I
Sbjct: 421 TNTLFGTKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEI------NKQQYSPSQIG 582
Query: 127 SMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAA 186
+ VLTKM+E AE +L + AVVTVPAYFND+QR+ATKDAG IAGL V RIINEPTAAA
Sbjct: 583 AFVLTKMKETAEAYLGKTISKAVVTVPAYFNDAQRQATKDAGRIAGLEVKRIINEPTAAA 762
Query: 187 LAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMV 246
L+YG+ + E I +FDLGGGTFDVS+L I N +FEVKAT+GDT LGGEDFDN ++
Sbjct: 763 LSYGMNNK-----EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALL 927
Query: 247 NHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID----FH 302
+ V EF R D++ L+RLR A E+ K LS + I++ + +
Sbjct: 928 DFLVSEFKRTDSIDLAKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLN 1107
Query: 303 SSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFF 362
++TR+KFE L +L ++ +SCL DA I VD+ +LVGG +R+PKVQ+++ E F
Sbjct: 1108ITLTRSKFEALVNNLIERTKAPCKSCLKDANISIKDVDEVLLVGGMTRVPKVQEVVSEIF 1287
Query: 363 NWKDICVSINPDEAVAYGAAVQAALLCEATEKSLNLVLRDVTPLSLGTLVRGDLLSVVIP 422
K +NPDEAVA GAA+Q +L ++ L+L DVTPLSLG G + + +I
Sbjct: 1288G-KSPSKGVNPDEAVAMGAALQGGILRGDVKE---LLLLDVTPLSLGIETLGGIFTRLIS 1455
Query: 423 RNTPIPVKKTKNYYTTKDDLSYVSVR--VYEGERLKASENNLLGKFRF-SIPPAPRGQIP 479
RNT IP KK++ + T D+ + EG R A++N LG+F IPPAPRG
Sbjct: 1456RNTTIPTKKSQVFSTAADNQTQRGYXRCSQEGXREMAADNKSLGEFDLVGIPPAPRGLPQ 1635
Query: 480 IKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKFK 539
I+V F ID +GI+ VSA + + G Q+ITI G LS +EI M++EAE D + K
Sbjct: 1636IEVTFDIDANGIVTVSAKDKSTGKEQQITI-RSSGGLSDDEINNMVKEAELHAQRDQERK 1812
Query: 540 KKVKAMNDLDDYLYTMRKVMK--------------DDSVSSMLTSIERMKINSAMIK 582
+ N D +Y++ K + ++SVS + T++E ++ K
Sbjct: 1813ALIDIKNSADTSIYSIEKSLSEYREKIPSEVAKEIENSVSDLRTAMEGESVDEIKTK 1983
>TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chaperone
HSC71.0 - garden pea, partial (59%)
Length = 1336
Score = 376 bits (965), Expect = e-104
Identities = 195/332 (58%), Positives = 248/332 (73%), Gaps = 2/332 (0%)
Frame = +2
Query: 266 RVLRRLRTACERTKRTLSFDTNATIDIDSLFEGIDFHSSITRAKFEQLNVDLFKKCIETV 325
R LRRLRTACER KRTLS TI+IDSLFEGIDF+S ITRA+FE+LN+DLF+KC+E V
Sbjct: 2 RALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFYSPITRARFEELNMDLFRKCMEPV 181
Query: 326 ESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNWKDICVSINPDEAVAYGAAVQA 385
E CL DA+++K SV D VLVGGS+RIPKVQQLL++FFN K++C SINPDEAVAYGAAVQA
Sbjct: 182 EKCLRDAKMDKKSVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQA 361
Query: 386 ALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSVVIPRNTPIPVKKTKNYYTTKDDLSY 444
A+L E EK +L+L DVTPLSLG G +++V+IPRNT IP KK + + T D+
Sbjct: 362 AILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPG 541
Query: 445 VSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQIPIKVCFSIDFDGILNVSATEDTCGN 503
V ++V+EGER + +NNLLGKF S IPPAPRG I VCF ID +GILNVSA + T G
Sbjct: 542 VLIQVFEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQ 721
Query: 504 RQEITITNEKGRLSTEEIERMIQEAENFKDEDMKFKKKVKAMNDLDDYLYTMRKVMKDDS 563
+ +ITITN+KGRLS E+IE+M+QEAE +K ED + KKKV+A N L++Y Y MR +KD+
Sbjct: 722 KNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTIKDEK 901
Query: 564 VSSMLTSIERMKINSAMIKGMKLIDEKQHHEA 595
++ L S ++ KI + ++ +D Q EA
Sbjct: 902 IAGKLDSDDKKKIEDTIEAAIQWLDANQLAEA 997
>TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (53%)
Length = 1189
Score = 311 bits (797), Expect = 5e-85
Identities = 160/292 (54%), Positives = 215/292 (72%), Gaps = 2/292 (0%)
Frame = +3
Query: 305 ITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNW 364
ITRA+FE+LN+DLF+KC+E VE CL DA+I+KS V + VLVGGS+RIPKVQQLL++FFN
Sbjct: 3 ITRARFEELNMDLFRKCMEPVEKCLRDAKIDKSHVHEVVLVGGSTRIPKVQQLLQDFFNG 182
Query: 365 KDICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSVVIPR 423
K++C SINPDEAVAYGAAVQAA+L E EK +L+L DVTPLSLG G +++ +IPR
Sbjct: 183 KELCKSINPDEAVAYGAAVQAAILSGEGDEKVQDLLLLDVTPLSLGLETAGGVMTTLIPR 362
Query: 424 NTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQIPIKV 482
NT IP KK + + T D+ V ++V+EGER + +NNLLGKF + IPPAPRG I V
Sbjct: 363 NTTIPTKKEQIFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELTGIPPAPRGVPQINV 542
Query: 483 CFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKFKKKV 542
CF ID +GILNVSA + T G + +ITITN+KGRLS EEIE+M+++AE +K ED + KKKV
Sbjct: 543 CFDIDANGILNVSAEDKTAGVKNKITITNDKGRLSKEEIEKMVKDAEKYKAEDEEVKKKV 722
Query: 543 KAMNDLDDYLYTMRKVMKDDSVSSMLTSIERMKINSAMIKGMKLIDEKQHHE 594
+A N +++Y Y MR +KD+ + L+ ++ KI A+ ++ ++ Q E
Sbjct: 723 EAKNSIENYAYNMRNTIKDEKIGGKLSHEDKEKIEKAVEDAIQWLEGNQMAE 878
>BQ139103 homologue to GP|2655420|gb| heat shock cognate protein HSC70
{Brassica napus}, partial (29%)
Length = 628
Score = 291 bits (746), Expect = 4e-79
Identities = 145/193 (75%), Positives = 167/193 (86%)
Frame = +2
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
MA K +GPAIGIDLGTTYSCV VWQ ++R EII NDQGNRTTPS+VAFTD +RLIGDAA
Sbjct: 56 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 229
Query: 61 KNQAASNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHF 120
KNQ A NP+NT+FDAKRLIGR++SD+ +Q+D++LWPFK++ G +KP I V YKG +K F
Sbjct: 230 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKIISGPAEKPLIGVNYKGEDKEF 409
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIIN 180
AEEISSMVL KMREIAE +L S +KNAVVTVPAYFNDSQR+ATKDAG IAGLNVMRIIN
Sbjct: 410 AAEEISSMVLMKMREIAEAYLGSAIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 589
Query: 181 EPTAAALAYGLQK 193
EPTAAA+AYGL K
Sbjct: 590 EPTAAAIAYGLDK 628
>TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic reticulum
HSC70-cognate binding protein precursor {Glycine max},
partial (38%)
Length = 962
Score = 288 bits (737), Expect = 4e-78
Identities = 152/230 (66%), Positives = 180/230 (78%), Gaps = 1/230 (0%)
Frame = +1
Query: 7 GPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII NDQGNR TPS+V+FTDD+RLIG+AAKN AA
Sbjct: 289 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 462
Query: 67 NPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGE-KHFCAEEI 125
NP TIFD KRLIGRK+ D +Q D++L P+K+V D KP I V+ K GE K F EEI
Sbjct: 463 NPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIV-NRDGKPYIQVRVKDGETKVFSPEEI 639
Query: 126 SSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAA 185
S+M+L KM+E AE FL +++AVVTVPAYFND+QR+ATKDAG IAGLNV RIINEPTAA
Sbjct: 640 SAMILGKMKETAEGFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 819
Query: 186 ALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTH 235
A+AYGL K+ E+NI +FDLGGGTFDVS+LTI N +FEV AT+GDTH
Sbjct: 820 AIAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTH 960
>TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chaperone BiP-A
- soybean, partial (52%)
Length = 1231
Score = 269 bits (688), Expect = 2e-72
Identities = 142/301 (47%), Positives = 210/301 (69%), Gaps = 3/301 (0%)
Frame = +1
Query: 297 EGIDFHSSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQ 356
+G+ F +TRA+FE+LN DLF+K + V+ + DA ++K+ +D+ VLVGGS+RIPKVQQ
Sbjct: 1 DGVAFSEPLTRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQ 180
Query: 357 LLKEFFNWKDICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGD 415
LLK++F+ K+ +NPDEAVA+GAAVQ ++L E ++ +++L DV PL+LG G
Sbjct: 181 LLKDYFDGKEPNKGVNPDEAVAFGAAVQGSILSGEGGAETKDILLLDVAPLTLGIETVGG 360
Query: 416 LLSVVIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAP 474
+++ +IPRNT IP KK++ + T +D + VS++V+EGER + LLGKF S IPPAP
Sbjct: 361 VMTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAP 540
Query: 475 RGQIPIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDE 534
RG I+V F +D +GILNV A + G ++ITITNEKGRLS EEIERM++EAE F +E
Sbjct: 541 RGTPQIEVTFEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIERMVREAEEFAEE 720
Query: 535 DMKFKKKVKAMNDLDDYLYTMRKVMKD-DSVSSMLTSIERMKINSAMIKGMKLIDEKQHH 593
D K K+++ A N L+ Y+Y M+ + D D ++ L S E+ KI +A+ + ++ +D+ Q
Sbjct: 721 DKKVKERIDARNALETYVYNMKNQVSDKDKLADKLESDEKEKIETAVKEALEWLDDNQSV 900
Query: 594 E 594
E
Sbjct: 901 E 903
>TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chaperone hsc70-3
- tomato, partial (27%)
Length = 667
Score = 264 bits (674), Expect = 8e-71
Identities = 135/194 (69%), Positives = 158/194 (80%)
Frame = +3
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
M+ + K AIGIDLGTTYSCVAV + N +II ND G RTTPSFVAF D +R+IGDAA
Sbjct: 81 MSGRGKKVAIGIDLGTTYSCVAVCK--NGEIDIIVNDLGKRTTPSFVAFKDSERMIGDAA 254
Query: 61 KNQAASNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHF 120
N AASNP NTIFDAKRLIGRK+SD ++Q+D++LWPFKV+ +DKP I+V Y EKHF
Sbjct: 255 FNIAASNPTNTIFDAKRLIGRKFSDPIVQSDVKLWPFKVIGDLNDKPMIVVNYNDEEKHF 434
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIIN 180
AEEISSMVL KMREIAE FL S V++ V+TVPAYFNDSQR++T+DAGAIAGLNVMRIIN
Sbjct: 435 AAEEISSMVLVKMREIAETFLGSIVEDVVITVPAYFNDSQRQSTRDAGAIAGLNVMRIIN 614
Query: 181 EPTAAALAYGLQKR 194
EPTAAA+AYG +
Sbjct: 615 EPTAAAIAYGFNTK 656
>TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock protein
{Arabidopsis thaliana}, partial (92%)
Length = 3027
Score = 247 bits (630), Expect = 1e-65
Identities = 147/446 (32%), Positives = 231/446 (50%), Gaps = 13/446 (2%)
Frame = +1
Query: 6 KGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAA 65
K +G D G VAV + + +++ ND+ R TP+ V F D +R IG A
Sbjct: 235 KMSVVGFDFGNESCIVAVARQRG--IDVVLNDESKRETPAIVCFGDKQRFIGTAGAASTM 408
Query: 66 SNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHFCAEEI 125
NP N+I KRLIG+K++D +Q D++ PF V G D P I +Y G + F A ++
Sbjct: 409 MNPKNSISQIKRLIGKKFADPELQRDLKSLPFNVTEGPDGYPLIHARYLGESREFTATQV 588
Query: 126 SSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAA 185
M+L+ ++EIA+K L + V + + +P YF D QR++ DA IAGL+ + +I+E TA
Sbjct: 589 FGMMLSNLKEIAQKNLNAAVVDCCIGIPVYFTDLQRRSVLDAATIAGLHPLHLIHETTAT 768
Query: 186 ALAYGLQKRANCVEE-RNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNR 244
ALAYG+ K E N+ D+G + V + K V + S D LGG DFD
Sbjct: 769 ALAYGIYKTDLPENEWLNVAFVDVGHASMQVCIAGFKKGQLHVLSHSYDRSLGGRDFDEA 948
Query: 245 MVNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGIDFHSS 304
+ +HF +F +Y D+ + R RLR ACE+ K+ LS + A ++I+ L + D
Sbjct: 949 LFHHFAAKFKEEYKIDVYQNARACLRLRAACEKLKKVLSANPEAPLNIECLMDEKDVRGF 1128
Query: 305 ITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNW 364
I R FEQL++ + ++ +E L++A + ++ +VG SR+P + ++L EFF
Sbjct: 1129IKRDDFEQLSLPILERVKGPLEKALAEAGLTVENIHMVEVVGSGSRVPAINKILTEFFK- 1305
Query: 365 KDICVSINPDEAVAYGAAVQAALLCEATEKSLNLVLRDVTPLSLGTLVR----------- 413
K+ ++N E VA GAA+Q A+L T K + + P S+ +
Sbjct: 1306KEPRRTMNASECVARGAALQCAIL-SPTFKVREFQVNESFPFSVSLSWKYSGSDAPDSES 1482
Query: 414 -GDLLSVVIPRNTPIPVKKTKNYYTT 438
++V P+ PIP K ++ T
Sbjct: 1483DNKQSTIVFPKGNPIPSSKVLTFFRT 1560
>AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 kDa protein.
[Petunia] {Petunia hybrida}, partial (24%)
Length = 532
Score = 243 bits (621), Expect = 1e-64
Identities = 120/167 (71%), Positives = 140/167 (82%)
Frame = +1
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
MA + A+GIDLGTTYSCVAVW ++ NR EIIHND+G++ TPSFVAFTD +RL+G AA
Sbjct: 31 MANNCECYAVGIDLGTTYSCVAVWLEEKNRVEIIHNDEGSKITPSFVAFTDGQRLVGAAA 210
Query: 61 KNQAASNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHF 120
K+QAA NP NT+FDAKRLIGRK+SD V+Q DI LWPFKVV G +DKP I +K+KG EK
Sbjct: 211 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDILLWPFKVVSGVNDKPMISLKFKGQEKLL 390
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDA 167
CAEEISS+VLT MREIAE +LESPVKNA +TVPAYFND+QRKAT DA
Sbjct: 391 CAEEISSIVLTNMREIAEMYLESPVKNAGITVPAYFNDAQRKATIDA 531
>TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [imported] -
Arabidopsis thaliana, partial (91%)
Length = 1717
Score = 139 bits (350), Expect(2) = 2e-60
Identities = 80/227 (35%), Positives = 125/227 (54%), Gaps = 3/227 (1%)
Frame = +1
Query: 271 LRTACERTKRTLSFDTNATIDIDSLFEGIDFHSSITRAKFEQLNVDLFKKCIETVESCLS 330
LR A + LS ++ +D+D L + + RA+FE++N ++F+KC + CL
Sbjct: 787 LRVATQDAIHQLSTQSSVQVDVD-LGNSLKICKVVDRAEFEEVNKEVFEKCESLIIQCLH 963
Query: 331 DAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNWKDICVSINPDEAVAYGAAVQAALLCE 390
DA++E ++D +LVGG S IP+V+ L+ ++ INP E GA ++ A+
Sbjct: 964 DAKVEVGDINDVILVGGCSYIPRVENLVTNLCKITEVYKGINPLEGAVCGATMEGAVASG 1143
Query: 391 ATEK--SLNLVLRDVTPLSLGTLVRGDLLSVVIPRNTPIPVKKTKNYYTTKDDLSYVSVR 448
++ +L+L+ TPL++G G+ VIPRNT +P +K + T D+ + +
Sbjct: 1144 ISDPFGNLDLLTIQATPLAIGIRADGNKFVPVIPRNTTMPARKDLLFTTIHDNQTEALIL 1323
Query: 449 VYEGERLKASENNLLGKFR-FSIPPAPRGQIPIKVCFSIDFDGILNV 494
VYEGE KA EN+LLG F+ IP AP+G I VC ID +L V
Sbjct: 1324 VYEGEGKKAEENHLLGYFKIMGIPAAPKGVPEISVCMDIDAANVLRV 1464
Score = 112 bits (279), Expect(2) = 2e-60
Identities = 80/245 (32%), Positives = 127/245 (51%), Gaps = 9/245 (3%)
Frame = +3
Query: 15 GTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAASNPV--NTI 72
GT+ VAVW + E++ N + + SFV F D+ G ++ + +TI
Sbjct: 3 GTSQCSVAVWN--GSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFGDTI 176
Query: 73 FDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDD-KPEIIVKYKGGEKHFCAEEISSMVLT 131
F+ KRLIGR +D V+ L PF V + +P I + EE+ +M L
Sbjct: 177 FNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAMFLV 353
Query: 132 KMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAALAYGL 191
++R + E L+ P++N V+TVP F+ Q + A A+AGL+V+R++ EPTA AL YG
Sbjct: 354 ELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALLYGQ 533
Query: 192 QKRANCVE------ERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRM 245
Q++ E E+ IF++G G DV++ + ++KA +G T +GGED M
Sbjct: 534 QQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDLLQNM 710
Query: 246 VNHFV 250
+ H +
Sbjct: 711 MRHLL 725
>BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated protein
homolog precursor (GRP 78), partial (47%)
Length = 1032
Score = 192 bits (487), Expect(3) = 1e-55
Identities = 100/215 (46%), Positives = 148/215 (68%), Gaps = 3/215 (1%)
Frame = -1
Query: 351 IPKVQQLLKEFFNWKDICVSINPDEAVAYGAAVQAALLC--EATEKSLNLVLRDVTPLSL 408
IPKVQ L++E+F K INPDEAVA+GAAVQA +L E TE+ +VL DV PL+L
Sbjct: 900 IPKVQSLIEEYFGGKKASKGINPDEAVAFGAAVQAGVLSGEEGTEE---IVLMDVNPLTL 730
Query: 409 GTLVRGDLLSVVIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRF 468
G G +++ +I RNTPIP +K++ + T D+ V ++V+EGER +NN LGKF
Sbjct: 729 GIETTGGVMTKLITRNTPIPTRKSQIFSTAADNQPVVLIQVFEGERSLTKDNNQLGKFEL 550
Query: 469 S-IPPAPRGQIPIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQE 527
+ IPPAPRG I+V F +D +GIL VSA + G ++ ITITN+KGRL+ EEI+RM++E
Sbjct: 549 TGIPPAPRGVPQIEVSFELDANGILKVSAHDKGTGKQESITITNDKGRLTKEEIDRMVEE 370
Query: 528 AENFKDEDMKFKKKVKAMNDLDDYLYTMRKVMKDD 562
AE + +ED +++++A N L++Y ++++ + D+
Sbjct: 369 AEKYAEEDKATRERIEARNGLENYAFSLKNQVNDE 265
Score = 38.5 bits (88), Expect(3) = 1e-55
Identities = 17/29 (58%), Positives = 22/29 (75%)
Frame = -3
Query: 325 VESCLSDAQIEKSSVDDAVLVGGSSRIPK 353
VE L DA+++K VDD VLVGGS+R P+
Sbjct: 979 VEQVLKDAKVKKEDVDDIVLVGGSTRYPQ 893
Score = 25.8 bits (55), Expect(3) = 1e-55
Identities = 10/16 (62%), Positives = 14/16 (87%)
Frame = -2
Query: 308 AKFEQLNVDLFKKCIE 323
AKFE+LN+DL KK ++
Sbjct: 1031 AKFEELNMDLLKKTLK 984
>TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protein 70
{Cucumis sativus}, partial (35%)
Length = 723
Score = 197 bits (501), Expect = 1e-50
Identities = 103/200 (51%), Positives = 139/200 (69%), Gaps = 1/200 (0%)
Frame = +3
Query: 397 NLVLRDVTPLSLGTLVRGDLLSVVIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLK 456
+L+L DVTPLSLG G +++V+IPRNT IP KK + + T D+ V ++VYEGER +
Sbjct: 6 DLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERAR 185
Query: 457 ASENNLLGKFRFS-IPPAPRGQIPIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGR 515
+NNLLGKF S IPPAPRG I VCF ID +GILNVSA + T G + +ITITN+KGR
Sbjct: 186 TRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGR 365
Query: 516 LSTEEIERMIQEAENFKDEDMKFKKKVKAMNDLDDYLYTMRKVMKDDSVSSMLTSIERMK 575
LS EEIE+M++EAE +K ED + KKKV A N L++Y Y MR +KD+ + S L+ ++ K
Sbjct: 366 LSKEEIEKMVEEAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIGSKLSPEDKKK 545
Query: 576 INSAMIKGMKLIDEKQHHEA 595
I+ A+ ++ +D Q EA
Sbjct: 546 IDDAIDAAIQWLDSNQLAEA 605
>AW695262 homologue to GP|170386|gb|A glucose-regulated protein 78
{Lycopersicon esculentum}, partial (39%)
Length = 607
Score = 177 bits (450), Expect = 8e-45
Identities = 96/158 (60%), Positives = 119/158 (74%), Gaps = 2/158 (1%)
Frame = +2
Query: 7 GPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII NDQGNR TPS+V+FTDD+RLIG+AAKN AA
Sbjct: 128 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 301
Query: 67 NPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGE-KHFCAEEI 125
NP TIFD KRLIGRK++D +Q D++L P+K+V D KP I V+ K GE K F EE+
Sbjct: 302 NPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIV-NKDGKPYIQVRVKDGETKVFSPEEV 478
Query: 126 SSMVLTKMREIAEKFLESPVKNAVVTVPAYF-NDSQRK 162
S+M+LTKM+E AE FL +++AVVTVPA ND+QR+
Sbjct: 479 SAMILTKMKETAEAFLGKTIRDAVVTVPALLSNDAQRQ 592
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,636,151
Number of Sequences: 36976
Number of extensions: 188640
Number of successful extensions: 897
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 824
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 827
length of query: 600
length of database: 9,014,727
effective HSP length: 102
effective length of query: 498
effective length of database: 5,243,175
effective search space: 2611101150
effective search space used: 2611101150
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144722.13


