

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.12 + phase: 0
(627 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
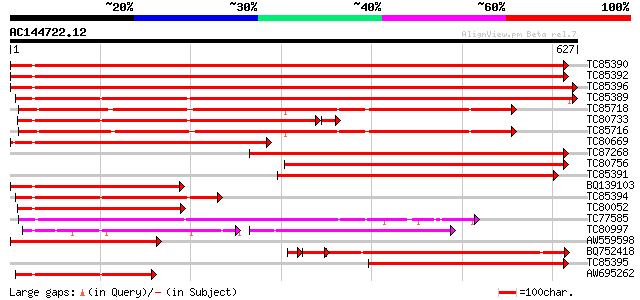
Score E
Sequences producing significant alignments: (bits) Value
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 806 0.0
TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chape... 805 0.0
TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protei... 798 0.0
TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protei... 630 0.0
TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa prot... 456 e-128
TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa prot... 431 e-125
TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chape... 434 e-122
TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 421 e-118
TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chape... 415 e-116
TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 348 4e-96
TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chape... 293 2e-79
BQ139103 homologue to GP|2655420|gb| heat shock cognate protein ... 280 9e-76
TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic retic... 275 4e-74
TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chapero... 256 2e-68
TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock... 254 9e-68
TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [im... 146 7e-60
AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 ... 226 2e-59
BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated pr... 204 4e-59
TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protei... 222 3e-58
AW695262 homologue to GP|170386|gb|A glucose-regulated protein 7... 166 3e-41
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 806 bits (2081), Expect = 0.0
Identities = 412/621 (66%), Positives = 500/621 (80%), Gaps = 3/621 (0%)
Frame = +1
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAA 60
MA K EGPAIGIDLGTTYSCV VWQ ++R EII N+QGNRTTPS+VAF+ +RLIGDAA
Sbjct: 136 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 309
Query: 61 KNQAASNPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHF 120
KNQ A NP NTVFDAKRLIGR++ D +++D++LWPFKVI G D P+IVV++K EEK F
Sbjct: 310 KNQVAMNPTNTVFDAKRLIGRRFGDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQF 489
Query: 121 VAEEISSMILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIIN 180
AEEISSM+L KMKEIAE +L + +K+AV+TVPAYFNDSQR+ATKDAGVISGLNV+RIIN
Sbjct: 490 SAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVLRIIN 669
Query: 181 EPTAAALAYGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGED 240
EPTAAA+AYGL K+A +N+ IFDLGGGTFDVS+LTI+ F+VKATAGDTHLGGED
Sbjct: 670 EPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 849
Query: 241 FDHRMVNHFVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEATIDIDAISEGID 300
FD+RMVNHFV+EFKRK+ KDISGN RALRRLRTACERAKRTLS + TI+ID++ EGID
Sbjct: 850 FDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGID 1029
Query: 301 FCSSITRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQD 360
F ++ITRA+FE+LNMDLF KCME VE+CL DA+MDK +V DVVLVGGS+RIPKV+QLLQD
Sbjct: 1030FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQD 1209
Query: 361 FFNGKDLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDVTPLSLGISIKGDVMSV 419
FFNGK+LC SINPDEAVAYGAAVQAA+L GEG + V +L+L DVTPLSLG+ G VM+V
Sbjct: 1210FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 1389
Query: 420 LIPRNSIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFELK-VPLAPRGLP 478
LIPRN+ IP KK+QV+ T D+QPGV + VYEGER+ +NNLLG FEL +P APRG+P
Sbjct: 1390LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVP 1569
Query: 479 -IQVCFAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAENFKEEDLKF 537
I VCF IDA+GILNVSAE++T+G K ITIT + GRLS EEIE+M+QEAE +K ED +
Sbjct: 1570QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEH 1749
Query: 538 KKKVKAMNVLDDYLYNMRKVMKDGSVTSMLTSTDKMKLNAAMIKGKNLIDDKEQHETFVF 597
KKKV+A N L++Y YNMR +KD + S L++ DK K+ A+ +D + E F
Sbjct: 1750KKKVEAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEADEF 1929
Query: 598 VDFLKELEKIFESALNKINKG 618
D +KELE I + ++ +G
Sbjct: 1930EDKMKELEGICNPIIARMYQG 1992
>TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chaperone
hsc70-3 - tomato, complete
Length = 2295
Score = 805 bits (2078), Expect = 0.0
Identities = 411/621 (66%), Positives = 501/621 (80%), Gaps = 3/621 (0%)
Frame = +1
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAA 60
MA K EGPAIGIDLGTTYSCV VWQ ++R EII N+QGNRTTPS+VAF+ +RLIGDAA
Sbjct: 130 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 303
Query: 61 KNQAASNPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHF 120
KNQ A NP NTVFDAKRLIGR+ SD +++D++LWPFKVI G D P+IVV++K EEK F
Sbjct: 304 KNQVAMNPTNTVFDAKRLIGRRISDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQF 483
Query: 121 VAEEISSMILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIIN 180
AEEISSM+L KMKEIAE +L + +K+AV+TVPAYFNDSQR+ATKDAGVISGLNV+RIIN
Sbjct: 484 SAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 663
Query: 181 EPTAAALAYGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGED 240
EPTAAA+AYGL K+A +N+ IFDLGGGTFDVS+LTI+ F+VKATAGDTHLGGED
Sbjct: 664 EPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 843
Query: 241 FDHRMVNHFVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEATIDIDAISEGID 300
FD+RMVNHFV+EFKRK+ KDISGN RALRR+RTACERAKRTLS + TI+ID++ EGID
Sbjct: 844 FDNRMVNHFVQEFKRKNKKDISGNPRALRRVRTACERAKRTLSSTAQTTIEIDSLFEGID 1023
Query: 301 FCSSITRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQD 360
F ++ITRA+FE+LNMDLF KCME VE+CL DA+MDK +VDDVVLVGGS+RIPKV+QLLQD
Sbjct: 1024FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVDDVVLVGGSTRIPKVQQLLQD 1203
Query: 361 FFNGKDLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDVTPLSLGISIKGDVMSV 419
FFNGK+LC SINPDEAVAYGAAVQAA+L GEG + V +L+L DVTPLSLG+ G VM+V
Sbjct: 1204FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 1383
Query: 420 LIPRNSIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFELK-VPLAPRGLP 478
LIPRN+ IP KK+QV+ T D+QPGV + VYEGER+ +NNLLG FEL +P APRG+P
Sbjct: 1384LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVP 1563
Query: 479 -IQVCFAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAENFKEEDLKF 537
I VCF IDA+GILNVSAE++T+G K ITIT + GRLS E+IE+M+QEAE +K ED +
Sbjct: 1564QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEH 1743
Query: 538 KKKVKAMNVLDDYLYNMRKVMKDGSVTSMLTSTDKMKLNAAMIKGKNLIDDKEQHETFVF 597
K+KV+A N L++Y YNMR +KD + S L++ DK K+ A+ +D + E F
Sbjct: 1744KRKVEAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEADEF 1923
Query: 598 VDFLKELEKIFESALNKINKG 618
D +KELE I + ++ +G
Sbjct: 1924EDKMKELEGICNPIIARMYQG 1986
>TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protein 70
{Cucumis sativus}, complete
Length = 2320
Score = 798 bits (2060), Expect = 0.0
Identities = 410/630 (65%), Positives = 503/630 (79%), Gaps = 3/630 (0%)
Frame = +2
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAA 60
MA K EGPAIGIDLGTTYSCV VWQ ++R EII N+QGNRTTPS+VAF+ +RLIGDAA
Sbjct: 107 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 280
Query: 61 KNQAASNPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHF 120
KNQ A NP NTVFDAKRLIGR+ SD+ +++D++LWPFKVIAG + P+I V++K EEK F
Sbjct: 281 KNQVAMNPTNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEEKLF 460
Query: 121 VAEEISSMILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIIN 180
+EEISSM+L KM+EIAE +L +K+AV+TVPAYFNDSQR+ATKDAGVI+GLNV+RIIN
Sbjct: 461 ASEEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 640
Query: 181 EPTAAALAYGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGED 240
EPTAAA+AYGL K+A V +N+ IFDLGGGTFDVS+LTI+ F+VKATAGDTHLGGED
Sbjct: 641 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 820
Query: 241 FDHRMVNHFVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEATIDIDAISEGID 300
FD+RMV HFV+EFKRK+ KDISGN RALRRLRTACERAKRTLS + TI+ID++ EG+D
Sbjct: 821 FDNRMVTHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVD 1000
Query: 301 FCSSITRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQD 360
F ++ITRA+FE+LNMDLF KCME VE+CL DA+MDK TV DVVLVGGS+RIPKV+QLLQD
Sbjct: 1001FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQD 1180
Query: 361 FFNGKDLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDVTPLSLGISIKGDVMSV 419
FFNGK+LC SINPDEAVAYGAAVQAA+L GEG + V +L+L DVTPLS G+ G VM+V
Sbjct: 1181FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSQGLETAGGVMTV 1360
Query: 420 LIPRNSIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFELK-VPLAPRGLP 478
LIPRN+ IP KK+QV+ T D+QPGV + VYEGER+ +NNLLG FEL +P APRG+P
Sbjct: 1361LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTKDNNLLGKFELSGIPPAPRGVP 1540
Query: 479 -IQVCFAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAENFKEEDLKF 537
I VCF IDA+GILNVSAE++T+G K ITIT + GRLS EEIE+M+QEAE +K ED +
Sbjct: 1541QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEH 1720
Query: 538 KKKVKAMNVLDDYLYNMRKVMKDGSVTSMLTSTDKMKLNAAMIKGKNLIDDKEQHETFVF 597
KKKV+A N L++Y YNMR +KD ++S L+ DK ++ A+ +D + E F
Sbjct: 1721KKKVEAKNSLENYAYNMRNTIKDEKISSKLSGGDKKQIEDAIEGAIQWLDANQLAEADEF 1900
Query: 598 VDFLKELEKIFESALNKINKGYSDEESDSD 627
D +KELE I + K+ +G + E + D
Sbjct: 1901EDKMKELETICNPIIAKMYQGGAGEGPEVD 1990
>TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protein 4
precursor (BiP 4) (78 kDa glucose-regulated protein
homolog 4) (GRP 78-4)., partial (94%)
Length = 2265
Score = 630 bits (1626), Expect = 0.0
Identities = 333/633 (52%), Positives = 449/633 (70%), Gaps = 12/633 (1%)
Frame = +2
Query: 7 GPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII N+QGNR TPS+V+F+ D+RLIG+AAKN AA
Sbjct: 155 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 328
Query: 67 NPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKE-EEKHFVAEEI 125
NP T+FD KRLIGRK++D ++ D++L P+K++ + P I V K+ E K F EE+
Sbjct: 329 NPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIV-NKDGKPYIQVRVKDGETKVFSPEEV 505
Query: 126 SSMILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIINEPTAA 185
S+MIL KMKE AE FL ++DAV+TVPAYFND+QR+ATKDAGVI+GLNV RIINEPTAA
Sbjct: 506 SAMILTKMKETAEAFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 685
Query: 186 ALAYGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGEDFDHRM 245
A+AYGL K+ +NI +FDLGGGTFDVS+LTI N F+V +T GDTHLGGEDFD R+
Sbjct: 686 AIAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLSTNGDTHLGGEDFDQRI 856
Query: 246 VNHFVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEATIDIDAISEGIDFCSSI 305
+ +F+K K+KHSKDIS ++RAL +LR ERAKR LS + ++I+++ +G+DF +
Sbjct: 857 MEYFIKLIKKKHSKDISKDNRALGKLRRESERAKRALSSQHQVRVEIESLFDGVDFSEPL 1036
Query: 306 TRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQDFFNGK 365
TRA+FE+LN DLF K M V++ + DA + K +D++VLVGGS+RIPKV+QLL+D+F+GK
Sbjct: 1037TRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFDGK 1216
Query: 366 DLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDVTPLSLGISIKGDVMSVLIPRN 424
+ +NPDEAVA+GAAVQ ++L GEG + +++L DV PL+LGI G VM+ LIPRN
Sbjct: 1217EPNKGVNPDEAVAFGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRN 1396
Query: 425 SIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFELK-VPLAPRGLP-IQVC 482
++IP KK QV+ T D Q VS+ V+EGERS+ + LLG F+L +P APRG P I+V
Sbjct: 1397TVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGNFDLSGIPPAPRGTPQIEVT 1576
Query: 483 FAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAENFKEEDLKFKKKVK 542
F +DA+GILNV AE++ +G + ITIT E GRLS EEI+RM++EAE F EED K K+++
Sbjct: 1577FEVDANGILNVRAEDKGTGKSEKITITNEKGRLSQEEIDRMVREAEEFAEEDKKVKERID 1756
Query: 543 AMNVLDDYLYNMRKVMKD-GSVTSMLTSTDKMKLNAAMIKGKNLIDDKEQHETFVFVDFL 601
A N L+ Y+YNM+ + D + L S +K K+ AA+ + +DD + E F + L
Sbjct: 1757ARNALETYVYNMKNQISDKDKLADKLESDEKEKIEAAVKEALEWLDDNQTVEKEEFEEKL 1936
Query: 602 KELEKIFESALNKINK-------GYSDEESDSD 627
KE+E + + + + G S E D D
Sbjct: 1937KEVEAVCNPIITAVYQRSGGAPGGASGEGEDED 2035
>TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa protein
mitochondrial precursor. [Kidney bean French bean]
{Phaseolus vulgaris}, complete
Length = 2371
Score = 456 bits (1172), Expect = e-128
Identities = 256/558 (45%), Positives = 367/558 (64%), Gaps = 7/558 (1%)
Frame = +2
Query: 10 IGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGD-QRLIGDAAKNQAASNP 68
IGIDLGTT SCV+V + +N + ++ N +G RTTPS VAF+ + L+G AK QA +NP
Sbjct: 263 IGIDLGTTNSCVSVMEGKNPK--VVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNP 436
Query: 69 ANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHFVAEEISSM 128
NT+ AKRLIGR++ D + ++++ P+K++ + + + + + + +I +
Sbjct: 437 ENTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWV----EAKGQQYSPSQIGAF 604
Query: 129 ILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIINEPTAAALA 188
+L KMKE AE +L V AVITVPAYFND+QR+ATKDAG I+GL V+RIINEPTAAAL+
Sbjct: 605 VLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALS 784
Query: 189 YGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGEDFDHRMVNH 248
YG+ K I +FDLGGGTFDVS+L I N F+VKAT GDT LGGEDFD+ +++
Sbjct: 785 YGMNKEGL------IAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDF 946
Query: 249 FVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEATIDIDAISEGIDFCS----S 304
V EFKR S D+S + AL+RLR A E+AK LS ++ I++ I+ +
Sbjct: 947 LVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNIT 1126
Query: 305 ITRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQDFFNG 364
+TR+KFE L +L E+ + CL DA + K +D+V+LVGG +R+PKV++++ F G
Sbjct: 1127LTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-G 1303
Query: 365 KDLCMSINPDEAVAYGAAVQAALLGEGIKSVPNLVLRDVTPLSLGISIKGDVMSVLIPRN 424
K C +NPDEAVA GAA+Q +L +K L+L DVTPLSLGI G + + LI RN
Sbjct: 1304KSPCKGVNPDEAVAMGAALQGGILRGDVK---ELLLLDVTPLSLGIETLGGIFTRLINRN 1474
Query: 425 SIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFEL-KVPLAPRGLP-IQVC 482
+ IP KK QV+ T D+Q V + V +GER +AS+N +LG FEL +P APRG+P I+V
Sbjct: 1475TTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVT 1654
Query: 483 FAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAENFKEEDLKFKKKVK 542
F IDA+GI+ VSA+++++G ++ ITI K +G LS +EI+ M++EAE ++D + K +
Sbjct: 1655FDIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLID 1831
Query: 543 AMNVLDDYLYNMRKVMKD 560
N D +Y++ K + +
Sbjct: 1832IRNSADTSIYSIEKSLSE 1885
>TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa protein
(HSP70). {Neurospora crassa}, partial (54%)
Length = 1258
Score = 431 bits (1107), Expect(2) = e-125
Identities = 221/335 (65%), Positives = 271/335 (79%)
Frame = +2
Query: 9 AIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAAKNQAASNP 68
AIGIDLGTTYSCV + N++ EII N+QGNRTTPS+VAF+ +RLIGDAAKNQ A NP
Sbjct: 188 AIGIDLGTTYSCVGRYA--NDKIEIIANDQGNRTTPSYVAFNDTERLIGDAAKNQVAMNP 361
Query: 69 ANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHFVAEEISSM 128
NTVFDAKRLIGRK+SDS ++ D++ +PFKVI P I V FK E K F EEIS+M
Sbjct: 362 HNTVFDAKRLIGRKFSDSEVQADMKHFPFKVI-DKGGKPNIEVEFKGENKTFTPEEISAM 538
Query: 129 ILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIINEPTAAALA 188
+L KM+E AE +L +V +AVITVPAYFNDSQR+ATKDAG+I+GLNV+RIINEPTAAA+A
Sbjct: 539 VLVKMRETAEAYLGGQVTNAVITVPAYFNDSQRQATKDAGLIAGLNVLRIINEPTAAAIA 718
Query: 189 YGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGEDFDHRMVNH 248
YGL K+A RN+ IFDLGGGTFDVS+LTI+ F+VK+TAGDTHLGGEDFD+R+VNH
Sbjct: 719 YGLDKKAE--GERNVLIFDLGGGTFDVSLLTIEEGIFEVKSTAGDTHLGGEDFDNRLVNH 892
Query: 249 FVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEATIDIDAISEGIDFCSSITRA 308
FV EFKRKH KD+S N+RALRRLRTACERAKRTLS + +I+ID++ EGIDF +SITRA
Sbjct: 893 FVNEFKRKHKKDLSSNARALRRLRTACERAKRTLSSSAQTSIEIDSLFEGIDFYTSITRA 1072
Query: 309 KFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVV 343
+FE+L DLF ++ V+R L+DA++DK V ++V
Sbjct: 1073RFEELCQDLFRSTIQPVDRVLSDAKIDKSQVHEIV 1177
Score = 37.0 bits (84), Expect(2) = e-125
Identities = 13/22 (59%), Positives = 21/22 (95%)
Frame = +3
Query: 345 VGGSSRIPKVKQLLQDFFNGKD 366
VGGS+RIP++++L+ D+FNGK+
Sbjct: 1182 VGGSTRIPRIQKLISDYFNGKE 1247
>TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chaperone PHSP1
precursor mitochondrial - garden pea, partial (98%)
Length = 2422
Score = 434 bits (1117), Expect = e-122
Identities = 248/560 (44%), Positives = 361/560 (64%), Gaps = 9/560 (1%)
Frame = +1
Query: 10 IGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGD-QRLIGDAAKNQAASNP 68
IGIDLGTT SCV++ + +N + +I N +G RTTPS VAF+ + L+G AK QA +NP
Sbjct: 247 IGIDLGTTNSCVSLMEGKNPK--VIENSEGARTTPSVVAFNQKGELLVGTPAKRQAVTNP 420
Query: 69 ANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHFVAEEISSM 128
NT+F KRLIGR++ D + ++++ P+K++ + + ++ ++ + +I +
Sbjct: 421 TNTLFGTKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEIN----KQQYSPSQIGAF 588
Query: 129 ILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIINEPTAAALA 188
+L KMKE AE +L + AV+TVPAYFND+QR+ATKDAG I+GL V RIINEPTAAAL+
Sbjct: 589 VLTKMKETAEAYLGKTISKAVVTVPAYFNDAQRQATKDAGRIAGLEVKRIINEPTAAALS 768
Query: 189 YGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGEDFDHRMVNH 248
YG+ + + +FDLGGGTFDVS+L I N F+VKAT GDT LGGEDFD+ +++
Sbjct: 769 YGMNNKEGLIA-----VFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDF 933
Query: 249 FVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEATIDIDAISEGIDFCS----S 304
V EFKR S D++ + AL+RLR A E+AK LS ++ I++ I+ +
Sbjct: 934 LVSEFKRTDSIDLAKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNIT 1113
Query: 305 ITRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQDFFNG 364
+TR+KFE L +L E+ + CL DA + K VD+V+LVGG +R+PKV++++ + F G
Sbjct: 1114LTRSKFEALVNNLIERTKAPCKSCLKDANISIKDVDEVLLVGGMTRVPKVQEVVSEIF-G 1290
Query: 365 KDLCMSINPDEAVAYGAAVQAALLGEGIKSVPNLVLRDVTPLSLGISIKGDVMSVLIPRN 424
K +NPDEAVA GAA+Q +L +K L+L DVTPLSLGI G + + LI RN
Sbjct: 1291KSPSKGVNPDEAVAMGAALQGGILRGDVK---ELLLLDVTPLSLGIETLGGIFTRLISRN 1461
Query: 425 SIIPVKKKQVYHTCDDDQP--GVSVDVYEGERSVASENNLLGLFEL-KVPLAPRGLP-IQ 480
+ IP KK QV+ T D+Q G EG R +A++N LG F+L +P APRGLP I+
Sbjct: 1462TTIPTKKSQVFSTAADNQTQRGYXRCSQEGXREMAADNKSLGEFDLVGIPPAPRGLPQIE 1641
Query: 481 VCFAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAENFKEEDLKFKKK 540
V F IDA+GI+ VSA+++++G ++ ITI + +G LS +EI M++EAE + D + K
Sbjct: 1642VTFDIDANGIVTVSAKDKSTGKEQQITI-RSSGGLSDDEINNMVKEAELHAQRDQERKAL 1818
Query: 541 VKAMNVLDDYLYNMRKVMKD 560
+ N D +Y++ K + +
Sbjct: 1819IDIKNSADTSIYSIEKSLSE 1878
>TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (44%)
Length = 955
Score = 421 bits (1081), Expect = e-118
Identities = 210/289 (72%), Positives = 246/289 (84%)
Frame = +2
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAA 60
MA K EG AIGIDLGTTYSCV VWQ N+R EII N+QGNRTTPS+VAF+ +RLIGDAA
Sbjct: 98 MATK-EGKAIGIDLGTTYSCVGVWQ--NDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAA 268
Query: 61 KNQAASNPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHF 120
KNQ A NP NTVFDAKRLIGR++SD ++ND++LWPFKV+ G + P+IVV++K EEK F
Sbjct: 269 KNQVAMNPQNTVFDAKRLIGRRFSDESVQNDMKLWPFKVVPGPAEKPMIVVNYKGEEKKF 448
Query: 121 VAEEISSMILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIIN 180
AEEISSM+L KM+E+AE FL VK+AV+TVPAYFNDSQR+ATKDAG ISGLNV+RIIN
Sbjct: 449 AAEEISSMVLIKMREVAEAFLGHPVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIIN 628
Query: 181 EPTAAALAYGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGED 240
EPTAAA+AYGL K+A+ +N+ IFDLGGGTFDVS+LTI+ F+VKATAGDTHLGGED
Sbjct: 629 EPTAAAIAYGLDKKASRKGEQNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 808
Query: 241 FDHRMVNHFVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEAT 289
FD+RMVNHFV EF+RK+ KDISGN+RALRRLRTACERAKRTLS + T
Sbjct: 809 FDNRMVNHFVSEFRRKNKKDISGNARALRRLRTACERAKRTLSSTAQTT 955
>TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chaperone
HSC71.0 - garden pea, partial (59%)
Length = 1336
Score = 415 bits (1066), Expect = e-116
Identities = 218/356 (61%), Positives = 270/356 (75%), Gaps = 3/356 (0%)
Frame = +2
Query: 266 RALRRLRTACERAKRTLSFDTEATIDIDAISEGIDFCSSITRAKFEQLNMDLFEKCMETV 325
RALRRLRTACERAKRTLS + TI+ID++ EGIDF S ITRA+FE+LNMDLF KCME V
Sbjct: 2 RALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFYSPITRARFEELNMDLFRKCMEPV 181
Query: 326 ERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQDFFNGKDLCMSINPDEAVAYGAAVQA 385
E+CL DA+MDKK+V DVVLVGGS+RIPKV+QLLQDFFNGK+LC SINPDEAVAYGAAVQA
Sbjct: 182 EKCLRDAKMDKKSVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQA 361
Query: 386 ALL-GEGIKSVPNLVLRDVTPLSLGISIKGDVMSVLIPRNSIIPVKKKQVYHTCDDDQPG 444
A+L GEG + V +L+L DVTPLSLG+ G VM+VLIPRN+ IP KK+QV+ T D+QPG
Sbjct: 362 AILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPG 541
Query: 445 VSVDVYEGERSVASENNLLGLFELK-VPLAPRGLP-IQVCFAIDADGILNVSAEEETSGN 502
V + V+EGER+ +NNLLG FEL +P APRG+P I VCF IDA+GILNVSAE++T+G
Sbjct: 542 VLIQVFEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQ 721
Query: 503 KKDITITKENGRLSTEEIERMIQEAENFKEEDLKFKKKVKAMNVLDDYLYNMRKVMKDGS 562
K ITIT + GRLS E+IE+M+QEAE +K ED + KKKV+A N L++Y YNMR +KD
Sbjct: 722 KNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTIKDEK 901
Query: 563 VTSMLTSTDKMKLNAAMIKGKNLIDDKEQHETFVFVDFLKELEKIFESALNKINKG 618
+ L S DK K+ + +D + E F D +KELE + + K+ +G
Sbjct: 902 IAGKLDSDDKKKIEDTIEAAIQWLDANQLAEADEFEDKMKELEGVCNPIIAKMYQG 1069
>TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (53%)
Length = 1189
Score = 348 bits (893), Expect = 4e-96
Identities = 183/317 (57%), Positives = 235/317 (73%), Gaps = 3/317 (0%)
Frame = +3
Query: 305 ITRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQDFFNG 364
ITRA+FE+LNMDLF KCME VE+CL DA++DK V +VVLVGGS+RIPKV+QLLQDFFNG
Sbjct: 3 ITRARFEELNMDLFRKCMEPVEKCLRDAKIDKSHVHEVVLVGGSTRIPKVQQLLQDFFNG 182
Query: 365 KDLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDVTPLSLGISIKGDVMSVLIPR 423
K+LC SINPDEAVAYGAAVQAA+L GEG + V +L+L DVTPLSLG+ G VM+ LIPR
Sbjct: 183 KELCKSINPDEAVAYGAAVQAAILSGEGDEKVQDLLLLDVTPLSLGLETAGGVMTTLIPR 362
Query: 424 NSIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFELK-VPLAPRGLP-IQV 481
N+ IP KK+Q++ T D+QPGV + V+EGER+ +NNLLG FEL +P APRG+P I V
Sbjct: 363 NTTIPTKKEQIFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELTGIPPAPRGVPQINV 542
Query: 482 CFAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAENFKEEDLKFKKKV 541
CF IDA+GILNVSAE++T+G K ITIT + GRLS EEIE+M+++AE +K ED + KKKV
Sbjct: 543 CFDIDANGILNVSAEDKTAGVKNKITITNDKGRLSKEEIEKMVKDAEKYKAEDEEVKKKV 722
Query: 542 KAMNVLDDYLYNMRKVMKDGSVTSMLTSTDKMKLNAAMIKGKNLIDDKEQHETFVFVDFL 601
+A N +++Y YNMR +KD + L+ DK K+ A+ ++ + E F D
Sbjct: 723 EAKNSIENYAYNMRNTIKDEKIGGKLSHEDKEKIEKAVEDAIQWLEGNQMAEVDEFEDKQ 902
Query: 602 KELEKIFESALNKINKG 618
KELE I + K+ +G
Sbjct: 903 KELEGICNPIIAKMYQG 953
>TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chaperone BiP-A
- soybean, partial (52%)
Length = 1231
Score = 293 bits (749), Expect = 2e-79
Identities = 154/315 (48%), Positives = 217/315 (68%), Gaps = 4/315 (1%)
Frame = +1
Query: 297 EGIDFCSSITRAKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQ 356
+G+ F +TRA+FE+LN DLF K M V++ + DA + K +D++VLVGGS+RIPKV+Q
Sbjct: 1 DGVAFSEPLTRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQ 180
Query: 357 LLQDFFNGKDLCMSINPDEAVAYGAAVQAALL-GEGIKSVPNLVLRDVTPLSLGISIKGD 415
LL+D+F+GK+ +NPDEAVA+GAAVQ ++L GEG +++L DV PL+LGI G
Sbjct: 181 LLKDYFDGKEPNKGVNPDEAVAFGAAVQGSILSGEGGAETKDILLLDVAPLTLGIETVGG 360
Query: 416 VMSVLIPRNSIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFELK-VPLAP 474
VM+ LIPRN++IP KK QV+ T D Q VS+ V+EGERS+ + LLG F+L +P AP
Sbjct: 361 VMTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAP 540
Query: 475 RGLP-IQVCFAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAENFKEE 533
RG P I+V F +DA+GILNV AE++ +G + ITIT E GRLS EEIERM++EAE F EE
Sbjct: 541 RGTPQIEVTFEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIERMVREAEEFAEE 720
Query: 534 DLKFKKKVKAMNVLDDYLYNMRKVMKD-GSVTSMLTSTDKMKLNAAMIKGKNLIDDKEQH 592
D K K+++ A N L+ Y+YNM+ + D + L S +K K+ A+ + +DD +
Sbjct: 721 DKKVKERIDARNALETYVYNMKNQVSDKDKLADKLESDEKEKIETAVKEALEWLDDNQSV 900
Query: 593 ETFVFVDFLKELEKI 607
E + + LKE+E +
Sbjct: 901 EKEEYEEKLKEVEAV 945
>BQ139103 homologue to GP|2655420|gb| heat shock cognate protein HSC70
{Brassica napus}, partial (29%)
Length = 628
Score = 280 bits (717), Expect = 9e-76
Identities = 137/193 (70%), Positives = 168/193 (86%)
Frame = +2
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAA 60
MA K EGPAIGIDLGTTYSCV VWQ ++R EII N+QGNRTTPS+VAF+ +RLIGDAA
Sbjct: 56 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 229
Query: 61 KNQAASNPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHF 120
KNQ A NP NTVFDAKRLIGR++SD+ +++D++LWPFK+I+G + P+I V++K E+K F
Sbjct: 230 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKIISGPAEKPLIGVNYKGEDKEF 409
Query: 121 VAEEISSMILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIIN 180
AEEISSM+L KM+EIAE +L S +K+AV+TVPAYFNDSQR+ATKDAGVI+GLNV+RIIN
Sbjct: 410 AAEEISSMVLMKMREIAEAYLGSAIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 589
Query: 181 EPTAAALAYGLQK 193
EPTAAA+AYGL K
Sbjct: 590 EPTAAAIAYGLDK 628
>TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic reticulum
HSC70-cognate binding protein precursor {Glycine max},
partial (38%)
Length = 962
Score = 275 bits (703), Expect = 4e-74
Identities = 143/230 (62%), Positives = 175/230 (75%), Gaps = 1/230 (0%)
Frame = +1
Query: 7 GPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII N+QGNR TPS+V+F+ D+RLIG+AAKN AA
Sbjct: 289 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 462
Query: 67 NPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKE-EEKHFVAEEI 125
NP T+FD KRLIGRK+ D ++ D++L P+K++ + P I V K+ E K F EEI
Sbjct: 463 NPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIV-NRDGKPYIQVRVKDGETKVFSPEEI 639
Query: 126 SSMILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIINEPTAA 185
S+MIL KMKE AE FL ++DAV+TVPAYFND+QR+ATKDAGVI+GLNV RIINEPTAA
Sbjct: 640 SAMILGKMKETAEGFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 819
Query: 186 ALAYGLQKRANCVENRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTH 235
A+AYGL K+ +NI +FDLGGGTFDVS+LTI N F+V AT GDTH
Sbjct: 820 AIAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTH 960
>TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chaperone hsc70-3
- tomato, partial (27%)
Length = 667
Score = 256 bits (654), Expect = 2e-68
Identities = 127/186 (68%), Positives = 154/186 (82%)
Frame = +3
Query: 9 AIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAAKNQAASNP 68
AIGIDLGTTYSCVAV + N +II N+ G RTTPSFVAF +R+IGDAA N AASNP
Sbjct: 105 AIGIDLGTTYSCVAVCK--NGEIDIIVNDLGKRTTPSFVAFKDSERMIGDAAFNIAASNP 278
Query: 69 ANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHFVAEEISSM 128
NT+FDAKRLIGRK+SD ++++D++LWPFKVI D P+IVV++ +EEKHF AEEISSM
Sbjct: 279 TNTIFDAKRLIGRKFSDPIVQSDVKLWPFKVIGDLNDKPMIVVNYNDEEKHFAAEEISSM 458
Query: 129 ILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIINEPTAAALA 188
+L KM+EIAE FL S V+D VITVPAYFNDSQR++T+DAG I+GLNV+RIINEPTAAA+A
Sbjct: 459 VLVKMREIAETFLGSIVEDVVITVPAYFNDSQRQSTRDAGAIAGLNVMRIINEPTAAAIA 638
Query: 189 YGLQKR 194
YG +
Sbjct: 639 YGFNTK 656
>TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock protein
{Arabidopsis thaliana}, partial (92%)
Length = 3027
Score = 254 bits (648), Expect = 9e-68
Identities = 164/531 (30%), Positives = 272/531 (50%), Gaps = 21/531 (3%)
Frame = +1
Query: 10 IGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAAKNQAASNPA 69
+G D G VAV +++ +++ N++ R TP+ V F QR IG A NP
Sbjct: 247 VGFDFGNESCIVAVARQRG--IDVVLNDESKRETPAIVCFGDKQRFIGTAGAASTMMNPK 420
Query: 70 NTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHFVAEEISSMI 129
N++ KRLIG+K++D ++ DL+ PF V G + P+I + E + F A ++ M+
Sbjct: 421 NSISQIKRLIGKKFADPELQRDLKSLPFNVTEGPDGYPLIHARYLGESREFTATQVFGMM 600
Query: 130 LAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIINEPTAAALAY 189
L+ +KEIA+K L + V D I +P YF D QR++ DA I+GL+ + +I+E TA ALAY
Sbjct: 601 LSNLKEIAQKNLNAAVVDCCIGIPVYFTDLQRRSVLDAATIAGLHPLHLIHETTATALAY 780
Query: 190 GLQKRANCVENR--NIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGEDFDHRMVN 247
G+ K + EN N+ D+G + V + K V + + D LGG DFD + +
Sbjct: 781 GIYK-TDLPENEWLNVAFVDVGHASMQVCIAGFKKGQLHVLSHSYDRSLGGRDFDEALFH 957
Query: 248 HFVKEFKRKHSKDISGNSRALRRLRTACERAKRTLSFDTEATIDIDAISEGIDFCSSITR 307
HF +FK ++ D+ N+RA RLR ACE+ K+ LS + EA ++I+ + + D I R
Sbjct: 958 HFAAKFKEEYKIDVYQNARACLRLRAACEKLKKVLSANPEAPLNIECLMDEKDVRGFIKR 1137
Query: 308 AKFEQLNMDLFEKCMETVERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQDFFNGKDL 367
FEQL++ + E+ +E+ L +A + + + V +VG SR+P + ++L +FF K+
Sbjct: 1138DDFEQLSLPILERVKGPLEKALAEAGLTVENIHMVEVVGSGSRVPAINKILTEFFK-KEP 1314
Query: 368 CMSINPDEAVAYGAAVQAALLGEGIKSVPNLVLRDVTPLSLGISIK------------GD 415
++N E VA GAA+Q A+L K V + + P S+ +S K
Sbjct: 1315RRTMNASECVARGAALQCAILSPTFK-VREFQVNESFPFSVSLSWKYSGSDAPDSESDNK 1491
Query: 416 VMSVLIPRNSIIPVKKKQVYHTCDDDQPGVSVDVY---EGERSVASENNLLGLFELKVPL 472
+++ P+ + IP K + SVDV E +G F+ K
Sbjct: 1492QSTIVFPKGNPIPSSKVLTFFRTGT----FSVDVQCHDLSETPTKISTYTIGPFQTK--N 1653
Query: 473 APRGLPIQVCFAIDADGILNVSAEEETSGNKKDITITK----ENGRLSTEE 519
+G ++ ++ GI++V + + ++ +TK EN ++ T+E
Sbjct: 1654GDKG-KVKAKVRLNLHGIVSVESATLFEEEEIEVPVTKEFAEENAKMETDE 1803
>TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [imported] -
Arabidopsis thaliana, partial (91%)
Length = 1717
Score = 146 bits (368), Expect(2) = 7e-60
Identities = 83/232 (35%), Positives = 134/232 (56%), Gaps = 4/232 (1%)
Frame = +1
Query: 266 RALRRLRTACERAKRTLSFDTEATIDIDAISEGIDFCSSITRAKFEQLNMDLFEKCMETV 325
+++ LR A + A LS + +D+D + + C + RA+FE++N ++FEKC +
Sbjct: 772 KSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKICKVVDRAEFEEVNKEVFEKCESLI 948
Query: 326 ERCLTDAEMDKKTVDDVVLVGGSSRIPKVKQLLQDFFNGKDLCMSINPDEAVAYGAAVQA 385
+CL DA+++ ++DV+LVGG S IP+V+ L+ + ++ INP E GA ++
Sbjct: 949 IQCLHDAKVEVGDINDVILVGGCSYIPRVENLVTNLCKITEVYKGINPLEGAVCGATMEG 1128
Query: 386 ALLGEGIKSVPNLVLRDV--TPLSLGISIKGDVMSVLIPRNSIIPVKKKQVYHTCDDDQP 443
A+ NL L + TPL++GI G+ +IPRN+ +P +K ++ T D+Q
Sbjct: 1129 AVASGISDPFGNLDLLTIQATPLAIGIRADGNKFVPVIPRNTTMPARKDLLFTTIHDNQT 1308
Query: 444 GVSVDVYEGERSVASENNLLGLFELK-VPLAPRGLP-IQVCFAIDADGILNV 493
+ VYEGE A EN+LLG F++ +P AP+G+P I VC IDA +L V
Sbjct: 1309 EALILVYEGEGKKAEENHLLGYFKIMGIPAAPKGVPEISVCMDIDAANVLRV 1464
Score = 103 bits (257), Expect(2) = 7e-60
Identities = 80/257 (31%), Positives = 126/257 (48%), Gaps = 16/257 (6%)
Frame = +3
Query: 15 GTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAAKNQAASNP---ANT 71
GT+ VAVW + E++ N++ + SFV F D+ G + + +T
Sbjct: 3 GTSQCSVAVWN--GSEVELLKNKRNQKLMKSFVTFK-DEAPSGGVTSQFSHEHEMLFGDT 173
Query: 72 VFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTED---NPVIVVHFKEEEKHFVAEEISSM 128
+F+ KRLIGR +D V+ L PF V T D P I + EE+ +M
Sbjct: 174 IFNMKRLIGRVDTDPVVHASKNL-PFLV--QTLDIGVRPFIAALVNNVWRSTTPEEVLAM 344
Query: 129 ILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDAGVISGLNVIRIINEPTAAALA 188
L +++ + E L+ +++ V+TVP F+ Q + A ++GL+V+R++ EPTA AL
Sbjct: 345 FLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALL 524
Query: 189 YGLQKRANCVE------NRNIFIFDLGGGTFDVSVLTIKNNAFDVKATAGDTHLGGEDFD 242
YG Q++ E + IF++G G DV+V +KA AG T +GGED
Sbjct: 525 YGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDLL 701
Query: 243 HRMVNHFVKE----FKR 255
M+ H + + FKR
Sbjct: 702 QNMMRHLLPDSENIFKR 752
>AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 kDa protein.
[Petunia] {Petunia hybrida}, partial (24%)
Length = 532
Score = 226 bits (576), Expect = 2e-59
Identities = 111/167 (66%), Positives = 135/167 (80%)
Frame = +1
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAA 60
MA E A+GIDLGTTYSCVAVW E+ NR EIIHN++G++ TPSFVAF+ QRL+G AA
Sbjct: 31 MANNCECYAVGIDLGTTYSCVAVWLEEKNRVEIIHNDEGSKITPSFVAFTDGQRLVGAAA 210
Query: 61 KNQAASNPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKEEEKHF 120
K+QAA NP NTVFDAKRLIGRK+SD V++ D+ LWPFKV++G D P+I + FK +EK
Sbjct: 211 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDILLWPFKVVSGVNDKPMISLKFKGQEKLL 390
Query: 121 VAEEISSMILAKMKEIAEKFLESKVKDAVITVPAYFNDSQRKATKDA 167
AEEISS++L M+EIAE +LES VK+A ITVPAYFND+QRKAT DA
Sbjct: 391 CAEEISSIVLTNMREIAEMYLESPVKNAGITVPAYFNDAQRKATIDA 531
>BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated protein
homolog precursor (GRP 78), partial (47%)
Length = 1032
Score = 204 bits (518), Expect(3) = 4e-59
Identities = 115/272 (42%), Positives = 175/272 (64%), Gaps = 3/272 (1%)
Frame = -1
Query: 351 IPKVKQLLQDFFNGKDLCMSINPDEAVAYGAAVQAALLGEGIKSVPNLVLRDVTPLSLGI 410
IPKV+ L++++F GK INPDEAVA+GAAVQA +L G + +VL DV PL+LGI
Sbjct: 900 IPKVQSLIEEYFGGKKASKGINPDEAVAFGAAVQAGVLS-GEEGTEEIVLMDVNPLTLGI 724
Query: 411 SIKGDVMSVLIPRNSIIPVKKKQVYHTCDDDQPGVSVDVYEGERSVASENNLLGLFELK- 469
G VM+ LI RN+ IP +K Q++ T D+QP V + V+EGERS+ +NN LG FEL
Sbjct: 723 ETTGGVMTKLITRNTPIPTRKSQIFSTAADNQPVVLIQVFEGERSLTKDNNQLGKFELTG 544
Query: 470 VPLAPRGLP-IQVCFAIDADGILNVSAEEETSGNKKDITITKENGRLSTEEIERMIQEAE 528
+P APRG+P I+V F +DA+GIL VSA ++ +G ++ ITIT + GRL+ EEI+RM++EAE
Sbjct: 543 IPPAPRGVPQIEVSFELDANGILKVSAHDKGTGKQESITITNDKGRLTKEEIDRMVEEAE 364
Query: 529 NFKEEDLKFKKKVKAMNVLDDYLYNMRKVMKD-GSVTSMLTSTDKMKLNAAMIKGKNLID 587
+ EED +++++A N L++Y ++++ + D + + DK + A+ + + ++
Sbjct: 363 KYAEEDKATRERIEARNGLENYAFSLKNQVNDEEGLGGKIDDEDKETILEAVKETNDWLE 184
Query: 588 DKEQHETFVFVDFLKELEKIFESALNKINKGY 619
E T DF ++ EK+ A +K Y
Sbjct: 183 --ENGATANTEDFEEQKEKLSNVAYPITSKMY 94
Score = 38.5 bits (88), Expect(3) = 4e-59
Identities = 17/29 (58%), Positives = 24/29 (82%)
Frame = -3
Query: 325 VERCLTDAEMDKKTVDDVVLVGGSSRIPK 353
VE+ L DA++ K+ VDD+VLVGGS+R P+
Sbjct: 979 VEQVLKDAKVKKEDVDDIVLVGGSTRYPQ 893
Score = 25.8 bits (55), Expect(3) = 4e-59
Identities = 10/16 (62%), Positives = 14/16 (87%)
Frame = -2
Query: 308 AKFEQLNMDLFEKCME 323
AKFE+LNMDL +K ++
Sbjct: 1031 AKFEELNMDLLKKTLK 984
>TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protein 70
{Cucumis sativus}, partial (35%)
Length = 723
Score = 222 bits (566), Expect = 3e-58
Identities = 118/224 (52%), Positives = 156/224 (68%), Gaps = 2/224 (0%)
Frame = +3
Query: 397 NLVLRDVTPLSLGISIKGDVMSVLIPRNSIIPVKKKQVYHTCDDDQPGVSVDVYEGERSV 456
+L+L DVTPLSLG+ G VM+VLIPRN+ IP KK+QV+ T D+QPGV + VYEGER+
Sbjct: 6 DLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERAR 185
Query: 457 ASENNLLGLFELK-VPLAPRGLP-IQVCFAIDADGILNVSAEEETSGNKKDITITKENGR 514
+NNLLG FEL +P APRG+P I VCF IDA+GILNVSAE++T+G K ITIT + GR
Sbjct: 186 TRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGR 365
Query: 515 LSTEEIERMIQEAENFKEEDLKFKKKVKAMNVLDDYLYNMRKVMKDGSVTSMLTSTDKMK 574
LS EEIE+M++EAE +K ED + KKKV A N L++Y YNMR +KD + S L+ DK K
Sbjct: 366 LSKEEIEKMVEEAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIGSKLSPEDKKK 545
Query: 575 LNAAMIKGKNLIDDKEQHETFVFVDFLKELEKIFESALNKINKG 618
++ A+ +D + E F D +KELE + + K+ +G
Sbjct: 546 IDDAIDAAIQWLDSNQLAEADEFQDKMKELESLCNPIIAKMYQG 677
>AW695262 homologue to GP|170386|gb|A glucose-regulated protein 78
{Lycopersicon esculentum}, partial (39%)
Length = 607
Score = 166 bits (419), Expect = 3e-41
Identities = 88/158 (55%), Positives = 115/158 (72%), Gaps = 2/158 (1%)
Frame = +2
Query: 7 GPAIGIDLGTTYSCVAVWQEQNNRAEIIHNEQGNRTTPSFVAFSGDQRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII N+QGNR TPS+V+F+ D+RLIG+AAKN AA
Sbjct: 128 GTVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 301
Query: 67 NPANTVFDAKRLIGRKYSDSVIKNDLQLWPFKVIAGTEDNPVIVVHFKE-EEKHFVAEEI 125
NP T+FD KRLIGRK++D ++ D++L P+K++ + P I V K+ E K F EE+
Sbjct: 302 NPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIV-NKDGKPYIQVRVKDGETKVFSPEEV 478
Query: 126 SSMILAKMKEIAEKFLESKVKDAVITVPAYF-NDSQRK 162
S+MIL KMKE AE FL ++DAV+TVPA ND+QR+
Sbjct: 479 SAMILTKMKETAEAFLGKTIRDAVVTVPALLSNDAQRQ 592
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.134 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,074,420
Number of Sequences: 36976
Number of extensions: 172290
Number of successful extensions: 904
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 830
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 833
length of query: 627
length of database: 9,014,727
effective HSP length: 102
effective length of query: 525
effective length of database: 5,243,175
effective search space: 2752666875
effective search space used: 2752666875
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144722.12


