

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144721.6 + phase: 0 /pseudo
(748 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
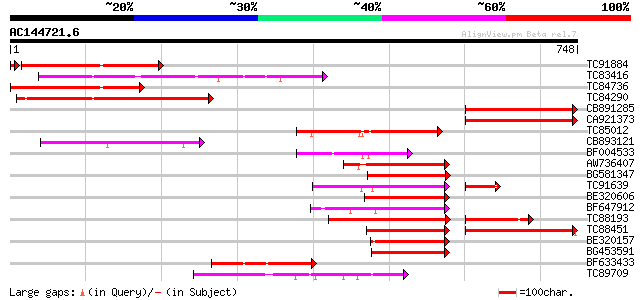
Score E
Sequences producing significant alignments: (bits) Value
TC91884 similar to GP|4741219|emb|CAB41879.1 SRK15 protein {Bras... 313 1e-86
TC83416 weakly similar to GP|3056594|gb|AAC13905.1| T1F9.15 {Ara... 301 8e-82
TC84736 299 3e-81
TC84290 weakly similar to GP|6554181|gb|AAF16627.1| T23J18.8 {Ar... 209 4e-54
CB891285 weakly similar to PIR|T14470|T14 receptor-like kinase (... 174 9e-44
CA921373 weakly similar to PIR|T14519|T14 probable S-receptor ki... 172 6e-43
TC85012 weakly similar to PIR|T05754|T05754 S-receptor kinase (E... 163 2e-40
CB893121 weakly similar to GP|4008008|gb|A receptor-like protein... 156 3e-38
BF004533 similar to PIR|H86246|H862 hypothetical protein [import... 149 4e-36
AW736407 similar to GP|4008008|gb| receptor-like protein kinase ... 149 4e-36
BG581347 similar to GP|13506745|gb receptor-like protein kinase ... 145 6e-35
TC91639 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1... 144 9e-35
BE320606 similar to GP|836954|gb|A receptor protein kinase {Ipom... 140 2e-33
BF647912 similar to GP|4530126|gb| receptor-like protein kinase ... 139 5e-33
TC88193 similar to PIR|G96602|G96602 probable receptor protein k... 137 2e-32
TC88451 similar to GP|836954|gb|AAC23542.1|| receptor protein ki... 136 3e-32
BE320157 weakly similar to PIR|T04835|T04 probable serine/threon... 135 4e-32
BG453591 similar to PIR|T05181|T05 S-receptor kinase (EC 2.7.1.-... 135 8e-32
BF633433 weakly similar to PIR|D96639|D966 protein T1F9.12 [impo... 134 1e-31
TC89709 weakly similar to GP|3868810|dbj|BAA34233.1 SRK23Bol {Br... 127 2e-29
>TC91884 similar to GP|4741219|emb|CAB41879.1 SRK15 protein {Brassica
oleracea}, partial (3%)
Length = 1156
Score = 313 bits (803), Expect(2) = 1e-86
Identities = 154/188 (81%), Positives = 169/188 (88%)
Frame = +3
Query: 16 FIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWYINET 75
F+IF TFYSCYS+ NDTITSSK LKDNETITSNNT+ KLGFFSPLNS NRYLGIWYINET
Sbjct: 585 FLIFCTFYSCYSAVNDTITSSKLLKDNETITSNNTDLKLGFFSPLNSPNRYLGIWYINET 764
Query: 76 NNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINSTAQLVDVGN 135
NNIWIANRDQPLKDSNGIVTIHKNGNLVILNK NGSIIWST+ISS S NSTA+L D GN
Sbjct: 765 NNIWIANRDQPLKDSNGIVTIHKNGNLVILNKPNGSIIWSTNISS--STNSTAKLDDAGN 938
Query: 136 LILSDINSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSGHYIGSLE 195
LIL DINS +TIWDSFTHP+D+AVP+M+IASNK TGK I+FV+RKS+NDPSSGH+ S+E
Sbjct: 939 LILRDINSGATIWDSFTHPSDSAVPSMKIASNKVTGKQIAFVARKSDNDPSSGHFTISVE 1118
Query: 196 RLDAPEVF 203
RLD PE F
Sbjct: 1119RLDVPESF 1142
Score = 25.4 bits (54), Expect(2) = 1e-86
Identities = 10/13 (76%), Positives = 10/13 (76%)
Frame = +2
Query: 1 MAFLIHNTNHFFI 13
MAFL HN NH FI
Sbjct: 542 MAFLSHNKNHLFI 580
>TC83416 weakly similar to GP|3056594|gb|AAC13905.1| T1F9.15 {Arabidopsis
thaliana}, partial (13%)
Length = 1142
Score = 301 bits (770), Expect = 8e-82
Identities = 159/390 (40%), Positives = 236/390 (59%), Gaps = 9/390 (2%)
Frame = +3
Query: 39 LKDNETITSNNTNFKLGFFSPLNSTNRYLGIWYINETNNIWIANRDQPL-KDSNGIVTIH 97
+KD+ET++SN+ NF LGFF+P NSTNRY+GIW + +W+ANR+QPL DS+G++ I
Sbjct: 12 IKDSETLSSNSGNFTLGFFTPQNSTNRYVGIWCKTQLFVVWVANRNQPLINDSSGVLEIS 191
Query: 98 KNGNLVILNKENGSIIWSTSISSPNSINSTAQLVDVGNLILSDINSRSTIWDSFTHPADA 157
+ N+V+LN + +++WST++++ ++ NS+ L D GNLIL + + TIW S HP ++
Sbjct: 192 NDNNIVLLNGKK-NVVWSTNLNNVSTSNSSVTLSDYGNLILFETTTEKTIWQSSDHPTNS 368
Query: 158 AVPTMRIASNKATGKNISFVSRKSENDPSSGHYIGSLERLDAPEVFIWYDKRIHWRTGPW 217
+P++ S N++ S K+ NDPS+G + +ERL+ PEVFI + R +WR+GPW
Sbjct: 369 ILPSLTFTS------NMTLTSWKTSNDPSNGSFTLGIERLNMPEVFIRKENRPYWRSGPW 530
Query: 218 NGTVFLGSPRMLTEYLAGWRFDQDKDGTTYLTYDFAVKA-MFGILSLTPNGTLKLVE--- 273
N +F+G M YL G+ F +D+ G T D +A +G++ N T ++ E
Sbjct: 531 NNQIFIGIEDMSALYLNGFHFQKDRTGG---TLDLVFRADDYGLVMYVVNSTGQMNEKSW 701
Query: 274 FLNNKEFLSLTVSQ-NECDFYGKCGPFGNCDISSVPNICSCFKGFEPKNLVEWSSRNWTN 332
+ N+E++ +Q ++CD YG CG FG C+ P IC C +GFEP+N EW+ +NWTN
Sbjct: 702 SIENEEWMDTWTNQRSDCDLYGFCGSFGICNSKGSP-ICRCLEGFEPRNNQEWNRQNWTN 878
Query: 333 GCVRKEGMNLKCEMVKNGSSVVK---QDKFLVHPNTKPPDFAERSDVSRDKCRTDCLANC 389
GCVRK L+CE N + D F+ K PDFAE V +D+C CL NC
Sbjct: 879 GCVRK--TPLQCESANNQNKSANGNDADSFMKLTLVKVPDFAELLSVEQDECENQCLMNC 1052
Query: 390 SCLAYAYDPFIRCMYWSSELIDLQKFPTSG 419
SC AY+Y I + ID+Q+F T G
Sbjct: 1053SCTAYSYVADIWLYVLEXKSIDIQQFETGG 1142
>TC84736
Length = 599
Score = 299 bits (765), Expect = 3e-81
Identities = 148/178 (83%), Positives = 160/178 (89%)
Frame = +1
Query: 1 MAFLIHNTNHFFIITFIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPL 60
MAFL HN+N+FFIITF+IF T YSC S+ NDTITSSKSLKDNETITSNNTNFKLG FSPL
Sbjct: 70 MAFLSHNSNYFFIITFLIFCTIYSCNSAINDTITSSKSLKDNETITSNNTNFKLGLFSPL 249
Query: 61 NSTNRYLGIWYINETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISS 120
NSTNRYLGIWYIN+TNNIWIANRDQPLKDSNGIVTIHK+GN +ILNK NG IIWST+ISS
Sbjct: 250 NSTNRYLGIWYINKTNNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISS 429
Query: 121 PNSINSTAQLVDVGNLILSDINSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVS 178
S NSTAQL D GNLIL DI+S +TIWDSFTHPADAAVPTMRIA+N+ TGK ISFVS
Sbjct: 430 --STNSTAQLADSGNLILRDISSGATIWDSFTHPADAAVPTMRIAANQVTGKKISFVS 597
>TC84290 weakly similar to GP|6554181|gb|AAF16627.1| T23J18.8 {Arabidopsis
thaliana}, partial (6%)
Length = 873
Score = 209 bits (531), Expect = 4e-54
Identities = 108/262 (41%), Positives = 163/262 (61%), Gaps = 2/262 (0%)
Frame = +1
Query: 10 HFFIITFIIFSTFYSC-YSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLG 68
+ F + FI+ YS +S DTITSS+ +KD ET+ S + NF LGFFSP NSTNRY+G
Sbjct: 97 NLFSVLFILCC--YSLQFSIAIDTITSSQFIKDPETLLSKDGNFTLGFFSPKNSTNRYVG 270
Query: 69 IWYINETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINSTA 128
IW+ +++ IW+ANR++ L DSNGI+TI ++GNL +LN + +IWS+++S+ S A
Sbjct: 271 IWWKSQSTIIWVANRNRSLNDSNGIITISEDGNLAVLNGQK-QVIWSSNLSNTTSNTIEA 447
Query: 129 QLVDVGNLILSDINSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSG 188
Q D GNL+L + + S +W SF P+D +P M++ SNK TG+ + S K+ +DPS G
Sbjct: 448 QFSDYGNLVLLESTTGSILWQSFQKPSDTLLPGMKLTSNKRTGEKVQLTSWKNPSDPSVG 627
Query: 189 HY-IGSLERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDGTTY 247
+ I ++R++ E+FI+ + + +WR+GPW+ VF G M T Y+ G R D +G Y
Sbjct: 628 SFSISFIDRINLHELFIFNETQPYWRSGPWDDVVFTGMQMMTTPYINGNRVGDDGEGNIY 807
Query: 248 LTYDFAVKAMFGILSLTPNGTL 269
+ Y + +L G L
Sbjct: 808 IYYTVPKDITMVVYNLNLQGHL 873
>CB891285 weakly similar to PIR|T14470|T14 receptor-like kinase (EC 2.7.1.-)
SFR2 - wild cabbage, partial (18%)
Length = 753
Score = 174 bits (442), Expect = 9e-44
Identities = 85/147 (57%), Positives = 112/147 (75%)
Frame = +2
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAM+GLFSEKSDV+SFGVLLLEI+SGRRNSSFY NE +L+L+GF W W E NI+S I
Sbjct: 194 PEYAMQGLFSEKSDVFSFGVLLLEIISGRRNSSFYDNEHALTLLGFVWIQWKEGNILSFI 373
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
+ E++D S ++RCIHIGLLCVQEL DRPN++ V+ ML SE LPPP + AF+ ++
Sbjct: 374 NTEIYDPSHHKYVVRCIHIGLLCVQELAVDRPNMAAVISMLNSEAELLPPPSQPAFILRQ 553
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
N S S ++S + S NSV+++++ G
Sbjct: 554 NMLSTMSHEESHRRYSINSVSITDISG 634
>CA921373 weakly similar to PIR|T14519|T14 probable S-receptor kinase (EC
2.7.1.-) SFR1 - wild cabbage, partial (13%)
Length = 779
Score = 172 bits (435), Expect = 6e-43
Identities = 84/147 (57%), Positives = 113/147 (76%)
Frame = -2
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAM GLFSEKSDV+SFGV LEIVSGRRNSSFY NE + SL+GF W W EEN++SLI
Sbjct: 763 PEYAMRGLFSEKSDVFSFGV*FLEIVSGRRNSSFYDNEHAPSLLGFVWIQWREENMLSLI 584
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D E++D S +++LRCIHIGLLCVQE DRP ++TV+ MLISE LPPP + AF+ ++
Sbjct: 583 DPEIYDHSHHTNILRCIHIGLLCVQESAVDRPTMATVISMLISEDAFLPPPSQPAFILRQ 404
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
N+ + +++Q S+N+V+++++ G
Sbjct: 403 NTLNSTQPEENQGVFSSNTVSITDMCG 323
>TC85012 weakly similar to PIR|T05754|T05754 S-receptor kinase (EC 2.7.1.-)
M4I22.110 precursor - Arabidopsis thaliana, partial
(17%)
Length = 645
Score = 163 bits (413), Expect = 2e-40
Identities = 89/217 (41%), Positives = 133/217 (61%), Gaps = 25/217 (11%)
Frame = +1
Query: 379 DKCRTDCLANCSCLAYAY----DPFIRCMYWSSELIDLQKFPTSGVDLFIRVPA-ELEKE 433
++C+T CL NCSC AYA D C+ W ++D++K P G D++IR+ + EL+ +
Sbjct: 1 EECKTMCLKNCSCTAYANSDIRDGGSGCLLWFDNIVDMRKHPDQGQDIYIRLASSELDHK 180
Query: 434 KGNKSFLIIAIAGGLGAFILVICAYLL----WRK----------------WSARHTEQKE 473
K + + G+ AFI+ + +L +RK W +H ++KE
Sbjct: 181 KNKRKLKLAGTLAGVVAFIIGLTVLVLITSVYRKKLGKPSENGYIKKLFLW--KHKKEKE 354
Query: 474 MKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIE 533
D ++DF + ATN+F + LG+GGFG VYKG++ DGQE+AVKRLSK+S QG E
Sbjct: 355 Y-CDLATIFDFSTITIATNNFSVKSKLGEGGFGAVYKGVMVDGQEIAVKRLSKTSAQGTE 531
Query: 534 EFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFM 570
EF NEV +++ LQHRNLV+LLGC +++ E++L+YEFM
Sbjct: 532 EFKNEVNLMATLQHRNLVKLLGCSIQQDEKLLIYEFM 642
>CB893121 weakly similar to GP|4008008|gb|A receptor-like protein kinase
{Arabidopsis thaliana}, partial (11%)
Length = 724
Score = 156 bits (394), Expect = 3e-38
Identities = 83/226 (36%), Positives = 129/226 (56%), Gaps = 10/226 (4%)
Frame = +1
Query: 41 DNETITSNNTNFKLGFFSPLNSTNRYLGIWYINETNN--IWIANRDQPLKDSNGIVTIHK 98
D E + S + F LGFF+P S +RY+GIWY N +W+ANRD P+ D++GI++I
Sbjct: 1 DGELLVSKSKTFALGFFTPGKSASRYVGIWYYNLPIQTVVWVANRDAPINDTSGILSIDP 180
Query: 99 NGNLVILNKENGSIIWSTSISSPNSI-NST----AQLVDVGNLILSDINSRSTIWDSFTH 153
NGNLVI + + IWST +S P S NST A+L D+ NL+L N+++ IW+SF H
Sbjct: 181 NGNLVIHHNHSTIPIWSTDVSFPQSQRNSTNAVIAKLSDIANLVLMINNTKTVIWESFDH 360
Query: 154 PADAAVPTMRIASNKATGKNISFVSRKSENDPSSGHYIGSLERLDAPEVFIWYDKRIHWR 213
P D +P ++I N+ T ++ S K+++DP G + + P++F++ WR
Sbjct: 361 PTDTLLPYLKIGFNRKTNQSWFLQSWKTDDDPGKGAFTVEFSTIGKPQLFMYNHNLPWWR 540
Query: 214 TGPWNGTVFLGSPRM---LTEYLAGWRFDQDKDGTTYLTYDFAVKA 256
G WNG +F G P M + + + D++ +Y +D +V A
Sbjct: 541 AGHWNGELFAGVPNMKRDMETFNVSFVEDENSVAISYNMFDKSVIA 678
>BF004533 similar to PIR|H86246|H862 hypothetical protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 571
Score = 149 bits (376), Expect = 4e-36
Identities = 83/189 (43%), Positives = 110/189 (57%), Gaps = 36/189 (19%)
Frame = +1
Query: 379 DKCRTDCLANCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLFIRVPAELEKEKGNKS 438
D CR CL NCSC AY+ D I CM W+ L+D+Q+ G+DL+ RV A E ++G
Sbjct: 7 DICRRLCLENCSCTAYSNDAGIGCMTWTGNLLDIQQLQMGGLDLYFRV-AHAELDRGGNK 183
Query: 439 FLIIAIAGGLGAFILVICAYLLWRK--------W----SARHTEQK-------------- 472
+II + +G I+ ICAY++WR+ W S R T +K
Sbjct: 184 TVIITTSVIIGTLIISICAYIMWRRTSNSSTKLWHSIKSTRKTNKKDFQLFNKGGTSDEN 363
Query: 473 ----------EMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVK 522
E++L EL L+DF KL ATN+FH SN LG+GGFGPVYKG L+DG+E+AVK
Sbjct: 364 NSDDVFGGLSEVRLQELLLFDFEKLATATNNFHLSNKLGQGGFGPVYKGKLQDGREIAVK 543
Query: 523 RLSKSSGQG 531
RLS++SGQG
Sbjct: 544 RLSRASGQG 570
>AW736407 similar to GP|4008008|gb| receptor-like protein kinase {Arabidopsis
thaliana}, partial (17%)
Length = 620
Score = 149 bits (376), Expect = 4e-36
Identities = 77/151 (50%), Positives = 103/151 (67%), Gaps = 11/151 (7%)
Frame = +2
Query: 441 IIAIAGGLGAFILVICAY----------LLWRKWSARHTEQKEMKLDELP-LYDFVKLEN 489
++A GL +LV Y LW+ +KE + EL ++DF + N
Sbjct: 32 VVAFIIGLNVLVLVTSVYRKKLGHIKKLFLWK-------HKKEKEDGELATIFDFSTITN 190
Query: 490 ATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRN 549
ATN+F N LG+GGFGPVYKG++ DGQE+AVKRLSK+SGQG EEF NEV +++ LQHRN
Sbjct: 191 ATNNFSVRNKLGEGGFGPVYKGVMVDGQEIAVKRLSKTSGQGTEEFKNEVKLMATLQHRN 370
Query: 550 LVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
LV+LLGC +++ E+ML+YEFMPN+SLD F+F
Sbjct: 371 LVKLLGCSIQQDEKMLIYEFMPNRSLDFFIF 463
>BG581347 similar to GP|13506745|gb receptor-like protein kinase 4
{Arabidopsis thaliana}, partial (15%)
Length = 826
Score = 145 bits (366), Expect = 6e-35
Identities = 70/109 (64%), Positives = 87/109 (79%)
Frame = +1
Query: 473 EMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGI 532
E+ E +DF + ATN+F N LG+GGFG VYKG+L +GQE+AVKRLS+SSGQGI
Sbjct: 319 EISRVEFLQFDFDTIATATNNFSGDNKLGEGGFGEVYKGMLFNGQEIAVKRLSRSSGQGI 498
Query: 533 EEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLFG 581
EEF NEV +++KLQHRNLVR+LG C++ E+ML+YEFMPNKSLD FLFG
Sbjct: 499 EEFKNEVVLVAKLQHRNLVRILGFCLDGEEKMLIYEFMPNKSLDYFLFG 645
>TC91639 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1.-)
T6K22.120 precursor - Arabidopsis thaliana, partial
(28%)
Length = 1313
Score = 144 bits (364), Expect = 9e-35
Identities = 87/216 (40%), Positives = 122/216 (56%), Gaps = 35/216 (16%)
Frame = +3
Query: 400 IRCMYWSSELIDLQKFPTS-GVDLFIRVP-AELEKEKGNKSFLIIAIAGGLGAFILV-IC 456
I CM W +L+D+ F G L IR+ ++L N+ +++ I L I + I
Sbjct: 21 IGCMVWYGDLVDILHFQHGEGNALHIRLAYSDLGDGGKNEKIMMVIILTSLAGLICIGII 200
Query: 457 AYLLWR-------------------------KWSARHTEQKEMKLD-------ELPLYDF 484
L+WR + SA E+ L+ ELP ++F
Sbjct: 201 VLLVWRYKRQLKASCSKNSDVLPVFDAHKSREMSAEIPGSVELGLEGNQLSKVELPFFNF 380
Query: 485 VKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEVAVISK 544
+ +ATN+F N LG+GGFGPVYKG L G+E+AVKRLS+ SGQG++EF NE+ + ++
Sbjct: 381 SCMSSATNNFSEENKLGQGGFGPVYKGKLPSGEEIAVKRLSRRSGQGLDEFKNEMRLFAQ 560
Query: 545 LQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
LQHRNLV+L+GC +E E++LVYEFM NKSLD FLF
Sbjct: 561 LQHRNLVKLMGCSIEGDEKLLVYEFMLNKSLDRFLF 668
Score = 73.6 bits (179), Expect = 3e-13
Identities = 37/46 (80%), Positives = 42/46 (90%)
Frame = +3
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGF 647
PEYAMEGL S KSDVYSFGVLLLEIVSGRRN+SF H++DS SL+G+
Sbjct: 921 PEYAMEGLVSVKSDVYSFGVLLLEIVSGRRNTSFRHSDDS-SLIGY 1055
>BE320606 similar to GP|836954|gb|A receptor protein kinase {Ipomoea
trifida}, partial (12%)
Length = 466
Score = 140 bits (353), Expect = 2e-33
Identities = 69/112 (61%), Positives = 86/112 (76%)
Frame = +1
Query: 469 TEQKEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSS 528
T +K M ELP +DF + ATN+F N LG+GGFG VYKG L +GQE+AVKRLSK+S
Sbjct: 76 TSEKNMDDLELPFFDFNTITMATNNFSEENKLGQGGFGIVYKGRLIEGQEIAVKRLSKNS 255
Query: 529 GQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
GQG++EF NEV +I KLQHRNLVRLLGC + E+MLVYE+M N+SLDA ++
Sbjct: 256 GQGVDEFKNEVRLIVKLQHRNLVRLLGCSFQMDEKMLVYEYMENRSLDAIVY 411
>BF647912 similar to GP|4530126|gb| receptor-like protein kinase homolog
RK20-1 {Phaseolus vulgaris}, partial (23%)
Length = 653
Score = 139 bits (349), Expect = 5e-33
Identities = 83/191 (43%), Positives = 114/191 (59%), Gaps = 8/191 (4%)
Frame = +3
Query: 398 PFIRCMYWSSELIDLQKFPTSGVDLFIRVPAELEKEKGNKSFLIIAIAGGL---GAFILV 454
P Y +S D PT +D P+E EK K + + + I + + A ++
Sbjct: 33 PSCNIRYETSSFYD----PTPAIDPGETSPSEEEKSKSSHTTIAIVVPTVVVVVAALLIF 200
Query: 455 ICAYLLWRKWSARHTEQKEMKLDELPL-----YDFVKLENATNSFHNSNMLGKGGFGPVY 509
IC L RK E +E D++ + ++F L+ AT++F +N LG GGFG VY
Sbjct: 201 ICTCLRKRKARINLEEIEEDDNDDIDMAESLQFNFETLQVATSNFSEANKLGHGGFGVVY 380
Query: 510 KGILEDGQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEF 569
GIL GQ +AVKRLS +SGQG EF NEV +++KLQHRNLVRLLG C+E E++LVYE+
Sbjct: 381 HGILAGGQVIAVKRLSTNSGQGDVEFKNEVLLVAKLQHRNLVRLLGFCLEGRERLLVYEY 560
Query: 570 MPNKSLDAFLF 580
+PNKSLD F+F
Sbjct: 561 VPNKSLDYFIF 593
>TC88193 similar to PIR|G96602|G96602 probable receptor protein kinase
F14G9.24 [imported] - Arabidopsis thaliana, partial
(20%)
Length = 1815
Score = 137 bits (345), Expect = 2e-32
Identities = 73/164 (44%), Positives = 106/164 (64%), Gaps = 3/164 (1%)
Frame = +3
Query: 421 DLFIRVPAELEKEKGNKSFLIIAIAGGLGA--FILVICAYLLWRKWSARHTEQKEMKLDE 478
D V ++ K NK LII + G+GA F+ V + + R+ + + + +D
Sbjct: 348 DFVPTVNSKPPSSKKNKVGLIIGVVVGVGAVCFLAVSSIFYILRRRKLYNDDDDLVGIDT 527
Query: 479 LP-LYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMN 537
+P + + +L+NAT+ F+ N LG+GGFGPVYKG L DG+ VAVK+LS S QG +F+
Sbjct: 528 MPNTFSYYELKNATSDFNRDNKLGEGGFGPVYKGTLNDGRFVAVKQLSIGSHQGKSQFIA 707
Query: 538 EVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLFG 581
E+A IS +QHRNLV+L GCC+E +++LVYE++ NKSLD LFG
Sbjct: 708 EIATISAVQHRNLVKLYGCCIEGNKRLLVYEYLENKSLDQALFG 839
Score = 65.5 bits (158), Expect = 7e-11
Identities = 36/89 (40%), Positives = 55/89 (61%)
Frame = +3
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAM G +EK+DV+SFGV+ LE+VSGR NS ED + L+ +AW+L I LI
Sbjct: 1080 PEYAMRGRLTEKADVFSFGVVALELVSGRPNSDSSLEEDKMYLLDWAWQLHERNCINDLI 1259
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPR 690
D + + + E + R + IG++ ++ +
Sbjct: 1260 DPRLSEFNME-EVERLVGIGIVVHSDITK 1343
Score = 31.2 bits (69), Expect = 1.5
Identities = 14/26 (53%), Positives = 18/26 (68%)
Frame = +1
Query: 681 GLLCVQELPRDRPNISTVVLMLISEI 706
GLLC Q P RP++S VV ML+ +I
Sbjct: 1315 GLLCTQTSPNLRPSMSRVVAMLLGDI 1392
>TC88451 similar to GP|836954|gb|AAC23542.1|| receptor protein kinase
{Ipomoea trifida}, partial (28%)
Length = 1276
Score = 136 bits (342), Expect = 3e-32
Identities = 62/110 (56%), Positives = 85/110 (76%)
Frame = +3
Query: 471 QKEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQ 530
+ + K ++ +++F + AT F N LG+GG+GPVYKGIL GQE+AVKRLSK+SGQ
Sbjct: 159 EDDFKGHDIKVFNFTSILEATMEFSPENKLGQGGYGPVYKGILATGQEIAVKRLSKTSGQ 338
Query: 531 GIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
GI EF NE+ +I +LQH+NLV+LLGCC+ E++L+YE+MPNKSLD +LF
Sbjct: 339 GIVEFKNELLLICELQHKNLVQLLGCCIHEEERILIYEYMPNKSLDFYLF 488
Score = 128 bits (321), Expect = 9e-30
Identities = 66/150 (44%), Positives = 97/150 (64%), Gaps = 3/150 (2%)
Frame = +3
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAMEG+ S KSDVYSFGVLLLEIV G +N+SFY + L+L+G AW+LW + + L+
Sbjct: 741 PEYAMEGVCSTKSDVYSFGVLLLEIVCGIKNNSFYDVDRPLNLIGHAWELWNDGEYLKLM 920
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D + D + RCIH+GLLCV++ DRP +S V+ +L ++ P K AF ++
Sbjct: 921 DPTLNDTFVPDEVKRCIHVGLLCVEQYANDRPTMSEVISVLTNKYVLTNLPRKPAFYVRR 1100
Query: 722 NSKSGESSQKSQQSNSNNSVTLS---EVQG 748
GE++ K Q +++ ++ T+S EV+G
Sbjct: 1101EIFEGETTSKGQDTDTYSTTTISTSFEVEG 1190
>BE320157 weakly similar to PIR|T04835|T04 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) F21P8.70 - Arabidopsis
thaliana, partial (27%)
Length = 529
Score = 135 bits (341), Expect = 4e-32
Identities = 66/105 (62%), Positives = 83/105 (78%)
Frame = +1
Query: 476 LDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEF 535
LD L + F +E ATN F + N +GKGGFG VYKG+L DGQ++AVK+LS+SSGQG EF
Sbjct: 7 LDSLQ-FKFSTIEAATNKFSSENEIGKGGFGIVYKGVLSDGQQIAVKKLSRSSGQGSIEF 183
Query: 536 MNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
NE+ +I+KLQHRNLV LLG C+E E+ML+YE++PNKSLD FLF
Sbjct: 184 QNEILLIAKLQHRNLVTLLGFCLEEREKMLIYEYVPNKSLDYFLF 318
>BG453591 similar to PIR|T05181|T05 S-receptor kinase (EC 2.7.1.-) T6K22.120
precursor - Arabidopsis thaliana, partial (24%)
Length = 640
Score = 135 bits (339), Expect = 8e-32
Identities = 65/104 (62%), Positives = 82/104 (78%), Gaps = 1/104 (0%)
Frame = +3
Query: 478 ELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILE-DGQEVAVKRLSKSSGQGIEEFM 536
ELP ++ + +ATN F N N LG+GGFGPVYKG L D +E+AVKRLS SS QG EF
Sbjct: 21 ELPFFNLSTIIDATNDFSNDNKLGEGGFGPVYKGTLVLDRREIAVKRLSGSSKQGTREFK 200
Query: 537 NEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
NEV + SKLQHRNLV++LGCC++ E+ML+YE+MPN+SLD+FLF
Sbjct: 201 NEVILCSKLQHRNLVKVLGCCIQGEEKMLIYEYMPNRSLDSFLF 332
Score = 33.5 bits (75), Expect = 0.31
Identities = 14/18 (77%), Positives = 17/18 (93%)
Frame = +3
Query: 602 PEYAMEGLFSEKSDVYSF 619
PEYA++GLFS KSDV+SF
Sbjct: 585 PEYAIDGLFSIKSDVFSF 638
>BF633433 weakly similar to PIR|D96639|D966 protein T1F9.12 [imported] -
Arabidopsis thaliana, partial (4%)
Length = 555
Score = 134 bits (338), Expect = 1e-31
Identities = 66/141 (46%), Positives = 92/141 (64%), Gaps = 2/141 (1%)
Frame = +3
Query: 267 GTLKLVEFLNNKEFLSLTVSQNECDFYGKCGPFGNCDISSVPNICSCFKGFEPKNLVEWS 326
G L ++N + + QNECD YG CG GNCD ++ P IC+C GFEP+N+ EW+
Sbjct: 123 GKLIYTSWMNKHQVGTTVAQQNECDIYGFCGLNGNCDSTNSP-ICTCLTGFEPRNVDEWN 299
Query: 327 SRNWTNGCVRKEGMNLKCEMVK-NGSSVV-KQDKFLVHPNTKPPDFAERSDVSRDKCRTD 384
+NW +GCVR+ +L+CE VK NGS + K+D F+ TK PDF ++S + D+C+T
Sbjct: 300 RQNWISGCVRRT--SLQCERVKYNGSELGGKEDGFVKLEMTKIPDFVQQSYLFVDECKTQ 473
Query: 385 CLANCSCLAYAYDPFIRCMYW 405
CL NC+C AYA+D IRC+ W
Sbjct: 474 CLNNCNCTAYAFDNGIRCLTW 536
>TC89709 weakly similar to GP|3868810|dbj|BAA34233.1 SRK23Bol {Brassica
oleracea}, partial (9%)
Length = 975
Score = 127 bits (319), Expect = 2e-29
Identities = 92/313 (29%), Positives = 147/313 (46%), Gaps = 29/313 (9%)
Frame = +2
Query: 243 DGTTYLTYDFAVKAMFGILSLTPNGTLKLVEFLN--NKEFLSLTVSQNECDFYGKCGPFG 300
D Y Y+ + I +L+P G L + + + + + T ++ CD Y CG
Sbjct: 50 DKEVYHWYELIDSSGVQIYALSPLGNLHALAWTSETSDRIVIETGMEDSCDSYAMCGANS 229
Query: 301 NCDISSVPNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVKQDKFL 360
C++ C C KG+ PK L +W+ W++GCV K +K+ FL
Sbjct: 230 VCNMDENVPKCECLKGYVPKFLEQWNISYWSSGCVPK---------IKSFCDNNSTTGFL 382
Query: 361 VHPNTKPPDFAERSD---VSRDKCRTDCLANCSCLAYAYDPFIR-----CMYWSSELIDL 412
+ K PD + D +S +C+ C+ NCSC AY + IR C+ W +LID+
Sbjct: 383 KYREMKLPDTSSSRDNKTMSLLECQKACMKNCSCTAYT-NADIRNGGSGCLLWIDDLIDM 559
Query: 413 QKFPTSGVDLFIRVPA-ELE--KEKGNKS----FLIIAIAGGLGAFILVICA-------- 457
+ F G DL+I+VP+ EL+ GNK+ + I ++ + F+ C
Sbjct: 560 RTFSQWGQDLYIKVPSSELDDVNVNGNKNQRQQLIRITVSVIISGFLTCACIIISIKIVA 739
Query: 458 ----YLLWRKWSARHTEQKEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGIL 513
+L +W ++ K+ + D LP++ F + AT +F + N LG+GGFGPVYKG L
Sbjct: 740 PRVYHLASFQWRKKYFRLKKEEPD-LPIFSFPIIVKATGNFSSXNKLGEGGFGPVYKGTL 916
Query: 514 EDGQEVAVKRLSK 526
G+ V + K
Sbjct: 917 IGGKXVXINGTQK 955
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.138 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,512,842
Number of Sequences: 36976
Number of extensions: 419666
Number of successful extensions: 3378
Number of sequences better than 10.0: 579
Number of HSP's better than 10.0 without gapping: 2952
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3133
length of query: 748
length of database: 9,014,727
effective HSP length: 103
effective length of query: 645
effective length of database: 5,206,199
effective search space: 3357998355
effective search space used: 3357998355
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144721.6


