

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144720.4 + phase: 0
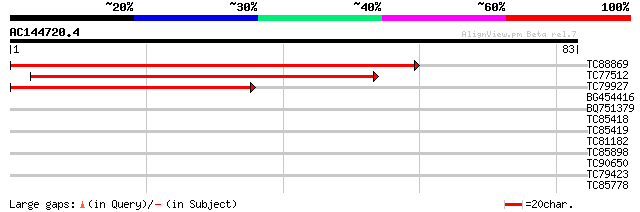
(83 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88869 similar to PIR|F84693|F84693 hypothetical protein At2g29... 103 2e-23
TC77512 similar to GP|22136912|gb|AAM91800.1 putative pumilio pr... 70 1e-13
TC79927 similar to PIR|F84693|F84693 hypothetical protein At2g29... 62 5e-11
BG454416 similar to GP|6056191|gb putative RNA binding protein {... 37 0.002
BQ751379 similar to GP|18376126|em conserved hypothetical protei... 35 0.004
TC85418 weakly similar to PIR|S23737|S23737 proline-rich protein... 26 3.1
TC85419 weakly similar to PIR|T05768|T05768 subtilisin-like prot... 26 3.1
TC81182 25 4.1
TC85898 similar to GP|12324950|gb|AAG52429.1 putative aminopepti... 25 5.3
TC90650 similar to GP|11994314|dbj|BAB02273. ankyrin-like protei... 25 7.0
TC79423 similar to GP|22136378|gb|AAM91267.1 HSR201-like protein... 25 7.0
TC85778 similar to GP|14517434|gb|AAK62607.1 AT4g01940/T7B11_20 ... 24 9.1
>TC88869 similar to PIR|F84693|F84693 hypothetical protein At2g29200
[imported] - Arabidopsis thaliana, partial (26%)
Length = 1217
Score = 103 bits (256), Expect = 2e-23
Identities = 49/60 (81%), Positives = 53/60 (87%)
Frame = +3
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTG 60
+QAMMKD F NYVVQKVLETCDD ELILSRIKVHLNALK+YTYGKHIV+RVEKL+ G
Sbjct: 603 LQAMMKDQFANYVVQKVLETCDDHQRELILSRIKVHLNALKKYTYGKHIVARVEKLVAAG 782
Score = 34.3 bits (77), Expect = 0.009
Identities = 14/49 (28%), Positives = 29/49 (58%)
Frame = +3
Query: 5 MKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
++D GN+V+QK +E +++++ I+S + L + YG ++ RV
Sbjct: 162 VRDQNGNHVIQKCIECVPEEAIDFIVSTFFDQVVTLSTHPYGCRVIQRV 308
Score = 28.9 bits (63), Expect = 0.37
Identities = 14/47 (29%), Positives = 27/47 (56%), Gaps = 1/47 (2%)
Frame = +3
Query: 8 PFGNYVVQKVLETCDDQSL-ELILSRIKVHLNALKRYTYGKHIVSRV 53
P+G V+Q+VLE C+ + + ++ I ++ L + YG ++V V
Sbjct: 279 PYGCRVIQRVLEHCESPATQQKVMDEILGAVSMLAQDQYGNYVVQHV 419
Score = 28.9 bits (63), Expect = 0.37
Identities = 16/58 (27%), Positives = 28/58 (47%)
Frame = +3
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGG 61
+ +D +GNYVVQ VLE I+ + + + + + ++ VEK +T G
Sbjct: 378 LAQDQYGNYVVQHVLEHGKPHERSTIIKELAGKIVQMSQQKFASNV---VEKCLTFSG 542
>TC77512 similar to GP|22136912|gb|AAM91800.1 putative pumilio protein
{Arabidopsis thaliana}, partial (37%)
Length = 1826
Score = 70.5 bits (171), Expect = 1e-13
Identities = 30/51 (58%), Positives = 39/51 (75%)
Frame = +3
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVE 54
MMKD F NYV+QKV++ C + +LS I+ H NALK+YTYGKHIV+R+E
Sbjct: 1167 MMKDQFANYVIQKVIDICSENQRARLLSHIRAHANALKKYTYGKHIVARME 1319
Score = 35.4 bits (80), Expect = 0.004
Identities = 15/50 (30%), Positives = 29/50 (58%)
Frame = +3
Query: 3 AMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSR 52
++ +D +GNYV Q VLE Q I+S++ H+ L ++ + ++V +
Sbjct: 930 SLAQDQYGNYVTQHVLERGRPQERSQIISKLSGHVVQLSQHKFASNVVEK 1079
Score = 33.5 bits (75), Expect = 0.015
Identities = 14/49 (28%), Positives = 27/49 (54%)
Frame = +3
Query: 5 MKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
++D GN+V+QK +E+ + ILS + + L + YG ++ R+
Sbjct: 717 VRDQNGNHVIQKCIESIPTNKIGFILSAFRGQVANLSMHPYGCRVIQRI 863
Score = 31.6 bits (70), Expect = 0.057
Identities = 16/72 (22%), Positives = 38/72 (52%)
Frame = +3
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNE 63
+M D FGNYV+QK E + + + + ++ + L YG ++ + ++I +
Sbjct: 498 LMTDVFGNYVIQKFFEYGNPEQRKELAEKLAGQILPLSLQMYGCRVIQKALEVI-EHEQK 674
Query: 64 CKLLRYMNSHLL 75
+L+R ++ +++
Sbjct: 675 AQLVRELDGNIM 710
Score = 29.6 bits (65), Expect = 0.22
Identities = 14/47 (29%), Positives = 26/47 (54%), Gaps = 1/47 (2%)
Frame = +3
Query: 8 PFGNYVVQKVLETCDDQ-SLELILSRIKVHLNALKRYTYGKHIVSRV 53
P+G V+Q++LE C D+ + I+ I + +L + YG ++ V
Sbjct: 834 PYGCRVIQRILEHCTDEVQCQFIVDEILESVCSLAQDQYGNYVTQHV 974
>TC79927 similar to PIR|F84693|F84693 hypothetical protein At2g29200
[imported] - Arabidopsis thaliana, partial (41%)
Length = 1844
Score = 61.6 bits (148), Expect = 5e-11
Identities = 30/36 (83%), Positives = 31/36 (85%)
Frame = +3
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVH 36
+QAMMKD F NYVVQKVLETCDDQ ELILSRI VH
Sbjct: 1737 LQAMMKDQFANYVVQKVLETCDDQQRELILSRI*VH 1844
Score = 33.9 bits (76), Expect = 0.011
Identities = 14/49 (28%), Positives = 28/49 (56%)
Frame = +3
Query: 5 MKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
++D GN+V+QK +E + +++ I+S + L + YG ++ RV
Sbjct: 1296 VRDQNGNHVIQKCIECVPEDAIDFIISTFFDQVVTLSTHPYGCRVIQRV 1442
Score = 33.5 bits (75), Expect = 0.015
Identities = 19/64 (29%), Positives = 33/64 (50%)
Frame = +3
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNE 63
+ +D +GNYVVQ VLE I+ + ++ + + + ++ VEK +T GG
Sbjct: 1512 LAQDQYGNYVVQHVLEHGKPHERSTIIKELAGNIVQMSQQKFASNV---VEKCLTFGGPS 1682
Query: 64 CKLL 67
+LL
Sbjct: 1683 ERLL 1694
Score = 32.3 bits (72), Expect = 0.033
Identities = 16/55 (29%), Positives = 28/55 (50%)
Frame = +3
Query: 3 AMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLI 57
A+M D FGNYVVQK E + +++ H+ L YG ++ + +++
Sbjct: 1074 ALMTDVFGNYVVQKFFEHGLASQRRELANKLYGHVLTLSLQMYGCRVIQKAIEVV 1238
Score = 29.3 bits (64), Expect = 0.28
Identities = 14/47 (29%), Positives = 28/47 (58%), Gaps = 1/47 (2%)
Frame = +3
Query: 8 PFGNYVVQKVLETCDD-QSLELILSRIKVHLNALKRYTYGKHIVSRV 53
P+G V+Q+VLE C++ + + ++ I ++ L + YG ++V V
Sbjct: 1413 PYGCRVIQRVLEHCENPDTQQKVMDEILGAVSMLAQDQYGNYVVQHV 1553
>BG454416 similar to GP|6056191|gb putative RNA binding protein {Arabidopsis
thaliana}, partial (20%)
Length = 657
Score = 36.6 bits (83), Expect = 0.002
Identities = 16/56 (28%), Positives = 31/56 (54%)
Frame = +3
Query: 2 QAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLI 57
+A+M D FGNYV+QK E D + + +++ H+ L YG ++ + +++
Sbjct: 198 RALMTDVFGNYVIQKFFEHGTDSQRKELANQLTGHVLPLSLQMYGCRVIQKALEVV 365
Score = 34.3 bits (77), Expect = 0.009
Identities = 16/38 (42%), Positives = 24/38 (63%)
Frame = +3
Query: 3 AMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNAL 40
A+ P+G V+Q+VLE CDD + I+ +HL+AL
Sbjct: 525 ALSTHPYGCRVIQRVLEHCDDLKTQEIIMEEIMHLSAL 638
Score = 33.9 bits (76), Expect = 0.011
Identities = 14/49 (28%), Positives = 28/49 (56%)
Frame = +3
Query: 5 MKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
++D GN+V+QK +E+ ++ I++ + AL + YG ++ RV
Sbjct: 423 VRDQNGNHVIQKCIESVPQNRIQFIITSFYGQVVALSTHPYGCRVIQRV 569
>BQ751379 similar to GP|18376126|em conserved hypothetical protein
{Neurospora crassa}, partial (30%)
Length = 771
Score = 35.4 bits (80), Expect = 0.004
Identities = 13/49 (26%), Positives = 29/49 (58%)
Frame = +1
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSR 52
+++DPFGNYVVQ +++ + E I+ + + L R+ + +++ +
Sbjct: 277 LVQDPFGNYVVQYIIDLNEPAFTEPIVRMFQSRIGQLSRHKFSSNVIEK 423
Score = 33.5 bits (75), Expect = 0.015
Identities = 14/54 (25%), Positives = 32/54 (58%)
Frame = +1
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVE 54
M+ +++D + NYV+Q LE +S ++ I+ L +++ YG+ I ++++
Sbjct: 490 MERLLRDSYANYVIQTALEYSTPRSKFRLVDVIRPILPSIRSTPYGRRIQAKIQ 651
>TC85418 weakly similar to PIR|S23737|S23737 proline-rich protein precursor
- kidney bean, partial (10%)
Length = 674
Score = 25.8 bits (55), Expect = 3.1
Identities = 11/28 (39%), Positives = 18/28 (64%)
Frame = -1
Query: 24 QSLELILSRIKVHLNALKRYTYGKHIVS 51
+SLE++ + +K N LK+YT K + S
Sbjct: 572 KSLEIVQAEVKPKKNCLKKYTKAKALQS 489
>TC85419 weakly similar to PIR|T05768|T05768 subtilisin-like proteinase (EC
3.4.21.-) - Arabidopsis thaliana, partial (23%)
Length = 879
Score = 25.8 bits (55), Expect = 3.1
Identities = 11/28 (39%), Positives = 18/28 (64%)
Frame = -1
Query: 24 QSLELILSRIKVHLNALKRYTYGKHIVS 51
+SLE++ + +K N LK+YT K + S
Sbjct: 831 KSLEIVQAEVKPKKNCLKKYTKAKALQS 748
>TC81182
Length = 917
Score = 25.4 bits (54), Expect = 4.1
Identities = 19/63 (30%), Positives = 25/63 (39%), Gaps = 4/63 (6%)
Frame = +2
Query: 21 CDDQSLELILS----RIKVHLNALKRYTYGKHIVSRVEKLITTGGNECKLLRYMNSHLLS 76
C QSL R H+ + R + + EK GGN+C LLR + L
Sbjct: 233 CQRQSLRTFTCKQT*RGTCHVMIITRVALSRRS*VKWEKTA*NGGNQC*LLRILTILSLL 412
Query: 77 LNC 79
NC
Sbjct: 413 PNC 421
>TC85898 similar to GP|12324950|gb|AAG52429.1 putative aminopeptidase;
4537-10989 {Arabidopsis thaliana}, partial (90%)
Length = 3374
Score = 25.0 bits (53), Expect = 5.3
Identities = 10/33 (30%), Positives = 20/33 (60%)
Frame = -3
Query: 47 KHIVSRVEKLITTGGNECKLLRYMNSHLLSLNC 79
++I++ +++ GG CKLL ++ S + L C
Sbjct: 2574 QNIITSFTRVLNYGGQCCKLLGHICSSFVFLKC 2476
>TC90650 similar to GP|11994314|dbj|BAB02273. ankyrin-like protein
{Arabidopsis thaliana}, partial (12%)
Length = 684
Score = 24.6 bits (52), Expect = 7.0
Identities = 13/42 (30%), Positives = 22/42 (51%), Gaps = 9/42 (21%)
Frame = +1
Query: 11 NYVVQKVLETCDDQSLELI---------LSRIKVHLNALKRY 43
N VV + + CDD+ ELI +R+K+ L+ ++ Y
Sbjct: 439 NIVVPRTIPVCDDRHSELIPCLDRNLIYQTRLKLDLSLMEHY 564
>TC79423 similar to GP|22136378|gb|AAM91267.1 HSR201-like protein
{Arabidopsis thaliana}, partial (7%)
Length = 848
Score = 24.6 bits (52), Expect = 7.0
Identities = 9/33 (27%), Positives = 19/33 (57%)
Frame = +1
Query: 8 PFGNYVVQKVLETCDDQSLELILSRIKVHLNAL 40
PF + K+ C+DQ + ++++IK L+ +
Sbjct: 256 PFAGRLKDKITIECNDQGVSFLVTKIKNKLSEI 354
>TC85778 similar to GP|14517434|gb|AAK62607.1 AT4g01940/T7B11_20
{Arabidopsis thaliana}, partial (50%)
Length = 555
Score = 24.3 bits (51), Expect = 9.1
Identities = 11/41 (26%), Positives = 22/41 (52%)
Frame = +3
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALK 41
++ ++K+ FG+ + V DDQ+ E + + HL L+
Sbjct: 378 IERVLKEKFGDSIEDIVQVFDDDQARETTVEAVNNHLEILR 500
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.136 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,771,765
Number of Sequences: 36976
Number of extensions: 33212
Number of successful extensions: 151
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 134
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 151
length of query: 83
length of database: 9,014,727
effective HSP length: 59
effective length of query: 24
effective length of database: 6,833,143
effective search space: 163995432
effective search space used: 163995432
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 51 (24.3 bits)
Medicago: description of AC144720.4


