

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144720.2 + phase: 0 /pseudo
(552 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
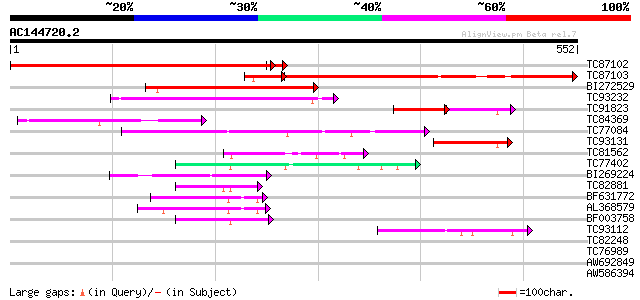
Score E
Sequences producing significant alignments: (bits) Value
TC87102 similar to PIR|T02589|T02589 hypothetical protein T16B24... 513 e-147
TC87103 similar to GP|15292735|gb|AAK92736.1 unknown protein {Ar... 394 e-124
BI272529 weakly similar to GP|6478925|gb| hypothetical protein {... 185 3e-47
TC93232 weakly similar to GP|21739227|emb|CAD40982. OSJNBa0072F1... 152 4e-37
TC91823 weakly similar to GP|19310729|gb|AAL85095.1 unknown prot... 53 1e-14
TC84369 similar to GP|19310729|gb|AAL85095.1 unknown protein {Ar... 74 1e-13
TC77084 similar to GP|15529155|gb|AAK97672.1 AT3g30390/T6J22_16 ... 65 9e-11
TC93131 similar to GP|19310729|gb|AAL85095.1 unknown protein {Ar... 59 6e-09
TC81562 weakly similar to GP|6671946|gb|AAF23206.1| putative ami... 49 4e-06
TC77402 similar to PIR|T50691|T50691 amino acid permease 6 [impo... 49 7e-06
BI269224 weakly similar to GP|11862972|db hypothetical protein {... 47 2e-05
TC82881 similar to GP|15216026|emb|CAC51423. amino acid permease... 46 4e-05
BF631772 similar to PIR|F86385|F86 hypothetical protein AAG12739... 45 7e-05
AL368579 similar to GP|14194151|gb AT5g40780/K1B16_3 {Arabidopsi... 43 4e-04
BF003758 similar to GP|15216030|em amino acid permease AAP4 {Vic... 43 4e-04
TC93112 weakly similar to GP|6671946|gb|AAF23206.1| putative ami... 42 6e-04
TC82248 similar to GP|15216028|emb|CAC51424. amino acid permease... 38 0.009
TC76989 homologue to SP|O24310|EFTU_PEA Elongation factor Tu ch... 35 0.075
AW692849 34 0.17
AW586394 similar to GP|15216026|em amino acid permease AAP1 {Vic... 33 0.37
>TC87102 similar to PIR|T02589|T02589 hypothetical protein T16B24.23 -
Arabidopsis thaliana (fragment), partial (44%)
Length = 964
Score = 513 bits (1322), Expect(2) = e-147
Identities = 256/258 (99%), Positives = 256/258 (99%)
Frame = +2
Query: 1 MNNSVASENSFIIESDEEDDKDFNKGDDGNDSDSSNYSNENPPQRKQSSYNPSWPQSYRQ 60
MNNSVASENSFIIESDEEDDKDFNKGDDGNDSDSSNYSNENPPQRKQSSYNPSWPQSYRQ
Sbjct: 155 MNNSVASENSFIIESDEEDDKDFNKGDDGNDSDSSNYSNENPPQRKQSSYNPSWPQSYRQ 334
Query: 61 SIDLYSSVPSPSIGFLGNSSLTRLSSSFLSTSLTRRHTPEVLPSVTKPLIQPTEEEKHQR 120
SIDLYSSVPSPSIGFLGNSSLTRLSSSFLSTSLTRRHTPEVLPSVTKPLIQPTEEEKHQR
Sbjct: 335 SIDLYSSVPSPSIGFLGNSSLTRLSSSFLSTSLTRRHTPEVLPSVTKPLIQPTEEEKHQR 514
Query: 121 RSSHTLLPPLSRRSSLLKKESKVSHEVPSRHCSFGQAVLNGINVLCGVGILSTPYAAKEG 180
RSSHTLLPPLSRRSSLLKKESKVSHEVPSRHCSFGQAVLNGINVLCGVGILSTPYAAKEG
Sbjct: 515 RSSHTLLPPLSRRSSLLKKESKVSHEVPSRHCSFGQAVLNGINVLCGVGILSTPYAAKEG 694
Query: 181 GWLGLSILFIFGILSFYTGLLLRSCLDSEPGLETYPDIGQAAFGTAGRIAISIVLYVELY 240
GWLGLSILFIFGILSFYTGLLLRSCLDSEPGLETYPDIGQAAFGTAGRIAISIVLYVELY
Sbjct: 695 GWLGLSILFIFGILSFYTGLLLRSCLDSEPGLETYPDIGQAAFGTAGRIAISIVLYVELY 874
Query: 241 GCCIEYIILEGDNLASLF 258
GCCIEYI LEGDNLA LF
Sbjct: 875 GCCIEYINLEGDNLAFLF 928
Score = 28.1 bits (61), Expect(2) = e-147
Identities = 14/20 (70%), Positives = 14/20 (70%)
Frame = +1
Query: 251 GDNLASLFPNAYLNLGGIEL 270
G L L NAYLNLGGIEL
Sbjct: 904 GRQLGFLVFNAYLNLGGIEL 963
>TC87103 similar to GP|15292735|gb|AAK92736.1 unknown protein {Arabidopsis
thaliana}, partial (51%)
Length = 1621
Score = 394 bits (1011), Expect(2) = e-124
Identities = 210/285 (73%), Positives = 218/285 (75%)
Frame = +3
Query: 268 IELNPQTLFAVVAALAVLPTVWLRDLSVLSYISAGGVIASVLVVLCLLWIGIEDVGFQRS 327
+ NPQTLFAVVAALAVLPTVWLRDLSVLSYISAGGVIASVLVVLCLLWIGIEDVGFQRS
Sbjct: 507 LNXNPQTLFAVVAALAVLPTVWLRDLSVLSYISAGGVIASVLVVLCLLWIGIEDVGFQRS 686
Query: 328 GTTLNLGTLPVAIGLYGYCYSGHAVFPNIYTSMAKPNQFPAVLVACFGVCTLLYAGGAVM 387
GTTLNLGTLPVAIGLYGYCYSGHAVFPNIYTSMAKPNQFPAVLVACFGVCTLLYAGGAVM
Sbjct: 687 GTTLNLGTLPVAIGLYGYCYSGHAVFPNIYTSMAKPNQFPAVLVACFGVCTLLYAGGAVM 866
Query: 388 GYKMFGEDTLSQFTLNLPQDLVATKIAVWTTVVNPFTKYALTISPVAMSLEELIPANHAK 447
GYK GEDTLSQFTLNLPQDLVAT + Y + I ++ E
Sbjct: 867 GYKNVGEDTLSQFTLNLPQDLVATXDCCLD--YGG*SIYQVCIDYISSGNES*RVDTSKP 1040
Query: 448 SYLFSIFIRTGLVFSTLVIGLSVPFFVALCYGKLAITCLIILCRFGDVLDWIITYNACNF 507
+ IF + P CY L+ L FGDVLDWIITYNACNF
Sbjct: 1041CQVLPIF----------HFHQNRPCIFYPCYWSLS-----SLFWFGDVLDWIITYNACNF 1175
Query: 508 DIAMCLLPKNLAGKSNANTGRTLHYNHCSRRCVFFRWNVLSPCGD 552
DIAMCLLPKNLAGKSNANTGRTLHYNHCSRRCVFFRWNVLSPCGD
Sbjct: 1176DIAMCLLPKNLAGKSNANTGRTLHYNHCSRRCVFFRWNVLSPCGD 1310
Score = 68.9 bits (167), Expect(2) = e-124
Identities = 34/43 (79%), Positives = 37/43 (85%), Gaps = 2/43 (4%)
Frame = +2
Query: 229 IAISIVLY--VELYGCCIEYIILEGDNLASLFPNAYLNLGGIE 269
+ IS+V Y V + GCCIEYIILEGDNLASLFPNAYLNLGGIE
Sbjct: 383 LTISVVHYNYVVMQGCCIEYIILEGDNLASLFPNAYLNLGGIE 511
Score = 35.4 bits (80), Expect = 0.057
Identities = 16/17 (94%), Positives = 17/17 (99%)
Frame = +1
Query: 217 DIGQAAFGTAGRIAISI 233
DIGQAAFGTAGRIAIS+
Sbjct: 10 DIGQAAFGTAGRIAISV 60
>BI272529 weakly similar to GP|6478925|gb| hypothetical protein {Arabidopsis
thaliana}, partial (23%)
Length = 625
Score = 185 bits (470), Expect = 3e-47
Identities = 90/179 (50%), Positives = 129/179 (71%), Gaps = 11/179 (6%)
Frame = +2
Query: 133 RSSLLKKESK----------VSHEVPS-RHCSFGQAVLNGINVLCGVGILSTPYAAKEGG 181
+SS L KE++ VS + + R CSF ++V+NG N+LCG+G+++ PYA KEGG
Sbjct: 41 KSSYLSKENQDDSNSLTIPFVSEKTSTPRQCSFARSVINGTNLLCGIGLMTMPYAVKEGG 220
Query: 182 WLGLSILFIFGILSFYTGLLLRSCLDSEPGLETYPDIGQAAFGTAGRIAISIVLYVELYG 241
+L L++L +F ++ YTG+LL CL S PGL+TYPDIGQAAFG AGR+ I+I+L +EL+G
Sbjct: 221 FLSLALLLLFAVICCYTGILLMRCLQSYPGLDTYPDIGQAAFGYAGRLGIAIILSLELFG 400
Query: 242 CCIEYIILEGDNLASLFPNAYLNLGGIELNPQTLFAVVAALAVLPTVWLRDLSVLSYIS 300
+E++ L DNL++LFPN + + G EL+ +F + AAL VLPT WL++LS+LSYIS
Sbjct: 401 ASVEFLTLVSDNLSALFPNTSMIVAGTELSTHQVFTIAAALLVLPTGWLKNLSLLSYIS 577
>TC93232 weakly similar to GP|21739227|emb|CAD40982. OSJNBa0072F16.6 {Oryza
sativa}, partial (56%)
Length = 695
Score = 152 bits (383), Expect = 4e-37
Identities = 81/224 (36%), Positives = 131/224 (58%), Gaps = 2/224 (0%)
Frame = +3
Query: 99 PEVLPSVTKPLIQPTEEEKHQRRSSHTLLPPLSRRSSLLKKESKVSHEVPSRHCSFGQAV 158
P LPS T+ + +E+ + + LL S+ +++ VSH SF
Sbjct: 6 PRFLPSTTQ-FLPMSEKVLNNSPLNVPLLSEWKLGHSIDEEKVIVSHPSNKNTVSFFHTC 182
Query: 159 LNGINVLCGVGILSTPYAAKEGGWLGLSILFIFGILSFYTGLLLRSCLDSEPGLETYPDI 218
+NG+N + GVGILS PYA GGWL L++LF +FY+G+L++ C++ ++TYPDI
Sbjct: 183 VNGLNAISGVGILSVPYALASGGWLSLALLFCIAAAAFYSGILMKRCMEKNSNIKTYPDI 362
Query: 219 GQAAFGTAGRIAISIVLYVELYGCCIEYIILEGDNLASLFPNAYLNLGGIELNPQTLFAV 278
G+ AFG GR+ +SI +Y ELY I ++ILEGDNL++LFP + G+ + + F +
Sbjct: 363 GELAFGKIGRLIVSISMYTELYLVSIGFLILEGDNLSNLFPIEEFQVFGLSIGAKKFFVI 542
Query: 279 VAALAVLPTVWLRDL--SVLSYISAGGVIASVLVVLCLLWIGIE 320
+ A+ +LPT+WL + + L Y+ V+ +LV+L + G++
Sbjct: 543 LVAVIILPTIWLDNFESAFLWYLQ---VVCLLLVLLFCQYHGLQ 665
>TC91823 weakly similar to GP|19310729|gb|AAL85095.1 unknown protein
{Arabidopsis thaliana}, partial (20%)
Length = 926
Score = 52.8 bits (125), Expect(2) = 1e-14
Identities = 24/55 (43%), Positives = 34/55 (61%)
Frame = +3
Query: 374 FGVCTLLYAGGAVMGYKMFGEDTLSQFTLNLPQDLVATKIAVWTTVVNPFTKYAL 428
F + LY G+ MFGE T SQ TL+LP+D A+K+++WT + P TK+ L
Sbjct: 48 FVLPVFLYGSVGAAGFLMFGERTSSQITLDLPRDAFASKVSLWTIAIIPLTKHML 212
Score = 44.7 bits (104), Expect(2) = 1e-14
Identities = 25/70 (35%), Positives = 42/70 (59%), Gaps = 3/70 (4%)
Frame = +2
Query: 426 YALTISPVAMSLEELIPANHAKSYLFSIFIRTGLVFSTLVIGLSVPFF---VALCYGKLA 482
YAL ++P+A SLEEL+P + +++ + +RT LV ST+ +PFF +AL L+
Sbjct: 206 YALMMNPLARSLEELLPDSISRTNWCFLLLRTALVISTVCAAFLIPFFGLVMALIGSLLS 385
Query: 483 ITCLIILCRF 492
+ ++L F
Sbjct: 386 VLVAMVLPAF 415
>TC84369 similar to GP|19310729|gb|AAL85095.1 unknown protein {Arabidopsis
thaliana}, partial (8%)
Length = 614
Score = 73.9 bits (180), Expect = 1e-13
Identities = 49/188 (26%), Positives = 89/188 (47%), Gaps = 4/188 (2%)
Frame = +3
Query: 8 ENSFIIESDEEDDKDFNKGDDGNDSDSSNYSNENPPQRKQSSYNPSWPQSYRQSIDLYSS 67
EN + ++++E D+ D + +DS ++ S ++ Q+ +S + WPQ+Y ++ID +
Sbjct: 27 ENEYFLDANE-DETDIEAVNYDSDSSNNRGSYDDDDQQPESFTSQQWPQTYNEAIDPLTI 203
Query: 68 VPSPSIGFLGNSSLTRLSS---SFLSTSLTRRHTPEVLPSVTKPLIQPTEEEKHQRRSSH 124
+P++G + SS SF S S H + + + ++ S
Sbjct: 204 AAAPNLGSILRGPSVLYSSFVGSFSSKSYLELHDGKTSFLSGNQIQEGFPTTWWEKASIQ 383
Query: 125 TLLPPLSRRSSLLKKESKVSHEVPSRH-CSFGQAVLNGINVLCGVGILSTPYAAKEGGWL 183
+P E+P + C+F Q + NGIN++ GVG+LSTP K+ GW+
Sbjct: 384 MQIP----------------EELPIGYGCTFTQTIFNGINIMAGVGLLSTPDTVKQAGWV 515
Query: 184 GLSILFIF 191
L ++ IF
Sbjct: 516 SLVVMLIF 539
>TC77084 similar to GP|15529155|gb|AAK97672.1 AT3g30390/T6J22_16
{Arabidopsis thaliana}, partial (88%)
Length = 2353
Score = 64.7 bits (156), Expect = 9e-11
Identities = 78/324 (24%), Positives = 133/324 (40%), Gaps = 25/324 (7%)
Frame = +1
Query: 110 IQPTEEEKHQRRSSHTLLPPLSRRSSLLKKESKVSHEVPSRH-CSFGQAVLNGINVLCGV 168
+ P E+K RSS +S + LL K + + SF AV N + G
Sbjct: 343 LAPKTEKKKSSRSSRKNKAVVSENAPLLPKSQESDSGFDDFNGASFSGAVFNLATTIIGA 522
Query: 169 GILSTPYAAKEGGWL-GLSILFIFGILSFYTGLLLRSCLDSEPGLETYPDIGQAAFGTAG 227
GI++ P K+ G + GL + + L+ + LL + + +Y + +FG G
Sbjct: 523 GIMALPATLKQLGLIPGLCAILLMAFLTEKSIELLIRFTRAGKAV-SYAGLMGDSFGKYG 699
Query: 228 RIAISIVLYVELYGCCIEYIILEGDNLASLFPNAYLNLGGIE-------LNPQTLFAVVA 280
+ I + V G I Y+I+ GD L+ + + G +E +T ++
Sbjct: 700 KAMAQICVIVNNIGVLIVYMIIIGDVLSGTSSSGEHHYGILEGWFGVHWWTGRTFVVLLT 879
Query: 281 ALAVL-PTVWLRDLSVLSYISAGGVIASVLVVLCLLWIGIEDVGFQRSGTTL-------- 331
+A+ P + + L + SA V ++ VV ++ +GI V G T+
Sbjct: 880 TVAIFTPLASFKRIDSLRFTSALSV--ALAVVFLVIAVGISIVKIISGGITMPRLFPAVT 1053
Query: 332 ------NLGTL-PVAIGLYGYCYSGHAVFPNIYTSMAKPNQFPAVLVACFGVCTLLYAGG 384
NL T+ PV + Y Y+ H+ I + +Q V+ G+C+ +Y
Sbjct: 1054DATSIVNLFTVVPVFVTAYICHYNVHS----IDNELEDNSQMQGVVRTALGLCSSVYLMI 1221
Query: 385 AVMGYKMFGEDTLSQFTLNLPQDL 408
+ G+ +FGE TL N DL
Sbjct: 1222SFFGFLLFGEGTLDDVLANFDADL 1293
>TC93131 similar to GP|19310729|gb|AAL85095.1 unknown protein {Arabidopsis
thaliana}, partial (22%)
Length = 786
Score = 58.5 bits (140), Expect = 6e-09
Identities = 31/80 (38%), Positives = 49/80 (60%), Gaps = 3/80 (3%)
Frame = +2
Query: 413 IAVWTTVVNPFTKYALTISPVAMSLEELIPANHAKSYLFSIFIRTGLVFSTLVIGLSVPF 472
+++WT V P TKYAL ++P+A S+EEL+P + + + I +RT LV ST+ +PF
Sbjct: 2 VSLWTIAVIPLTKYALMMNPLARSVEELLPHSISSTNWCFILLRTALVISTVCAAFVIPF 181
Query: 473 F---VALCYGKLAITCLIIL 489
F +AL L++ I+L
Sbjct: 182 FGLVMALIGSLLSVLVAIVL 241
>TC81562 weakly similar to GP|6671946|gb|AAF23206.1| putative amino acid
transporter protein {Arabidopsis thaliana}, partial
(36%)
Length = 673
Score = 49.3 bits (116), Expect = 4e-06
Identities = 42/150 (28%), Positives = 69/150 (46%), Gaps = 9/150 (6%)
Frame = +1
Query: 209 EPGLET--YPDIGQAAFGTAGRIAISIVLYVELYGCCIEYIILEGDNLASLFPNAYLNLG 266
EP +E+ Y D+G +G GR ++ V I Y++ G NL S+F +L
Sbjct: 226 EPLVESRSYGDLGYRCYGILGRYLTEFIIVVAQCAGAIAYLVFIGQNLDSVFQKYGPSLA 405
Query: 267 GIELNPQTLFAVVAALAVLPTVWLRDLSVLS----YISAGGVIASVLVVLCLLWIGIEDV 322
+F +V + W+ LS L+ + V+A +VV + + +ED
Sbjct: 406 SY------IFMLVPV--EIGMSWIGSLSALAPFSIFADVCNVLAMGIVVKEDVQVALED- 558
Query: 323 GF---QRSGTTLNLGTLPVAIGLYGYCYSG 349
GF +R+ T N+G LP A G+ +C+ G
Sbjct: 559 GFSFRERTAITSNIGGLPFAAGMAVFCFEG 648
>TC77402 similar to PIR|T50691|T50691 amino acid permease 6 [imported] -
Arabidopsis thaliana, partial (90%)
Length = 1902
Score = 48.5 bits (114), Expect = 7e-06
Identities = 57/270 (21%), Positives = 103/270 (38%), Gaps = 31/270 (11%)
Frame = +3
Query: 162 INVLCGVGILSTPYAAKEGGWL-GLSILFIFGILSFYTGLLLRSCLDSEPGLE-----TY 215
I + G G+LS +A + GW+ G ++LF F ++++T LL C S + TY
Sbjct: 396 ITAVIGSGVLSLAWAIAQMGWVAGPAVLFAFSFITYFTSTLLADCYRSPDPVHGKRNYTY 575
Query: 216 PDIGQAAFGTAGRIAISIVLYVELYGCCIEYIILEGDNLASLFPNAYLNLGG------IE 269
++ +A G + Y+ L G I Y I ++ ++ + + G +
Sbjct: 576 TEVVRANLGGRKFQLCGLAQYINLVGVTIGYTITASISMVAVQRSNCFHKHGHQDKCYVS 755
Query: 270 LNPQTLFAVVAALAVLPTVWLRDLSVLSYISAGGVIASVLVVLCLLWIGIEDVGFQRSGT 329
NP F ++ A + + + LS++S + S L + + V +
Sbjct: 756 NNP---FMIIFACIQIVLCQIPNFHELSWLSIVAAVMSFAYSSIGLGLSVAKVAGGGNHV 926
Query: 330 TLNLGTLPV---------------AIGLYGYCYSGHAVFPNIYTSM--AKPNQFPAVLVA 372
T +L + + AIG + Y+ V I ++ + P +
Sbjct: 927 TTSLTGVQIGVDVTATEKVWRMFQAIGDIAFAYAFSNVLIEIQDTLKSSPPENRVMKRAS 1106
Query: 373 CFGV--CTLLYAGGAVMGYKMFGEDTLSQF 400
G+ TL Y +GY FG D F
Sbjct: 1107LIGILTTTLFYVLCGTLGYAAFGNDAPGNF 1196
>BI269224 weakly similar to GP|11862972|db hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (26%)
Length = 537
Score = 47.0 bits (110), Expect = 2e-05
Identities = 40/159 (25%), Positives = 69/159 (43%), Gaps = 1/159 (0%)
Frame = +2
Query: 98 TPEVLPSVTKPLIQPTEEEKHQRRSSHTLLPPLSRRSSLLKKESKVSHEVPSRHCSFGQA 157
T E++ PL+Q +EK + H + S++ SF +
Sbjct: 17 TNEIVGEQYCPLLQTKTDEKIEVEIEHG-------------DDGSNSNKNNESAASFSGS 157
Query: 158 VLNGINVLCGVGILSTPYAAKEGGW-LGLSILFIFGILSFYTGLLLRSCLDSEPGLETYP 216
V N + G GI++ P A K G +G++ + +LS +T L + ++Y
Sbjct: 158 VFNLSTTIIGAGIMALPAAMKVLGLTIGIASIIFLALLS-HTSLDILMRFSRVAKAQSYG 334
Query: 217 DIGQAAFGTAGRIAISIVLYVELYGCCIEYIILEGDNLA 255
D+ AFG+ GR+ I + +G + YII+ GD L+
Sbjct: 335 DVMGYAFGSLGRLLFQISVLFNNFGILVVYIIIIGDVLS 451
>TC82881 similar to GP|15216026|emb|CAC51423. amino acid permease AAP1
{Vicia faba var. minor}, partial (28%)
Length = 1132
Score = 45.8 bits (107), Expect = 4e-05
Identities = 29/91 (31%), Positives = 42/91 (45%), Gaps = 6/91 (6%)
Frame = +2
Query: 162 INVLCGVGILSTPYAAKEGGW-LGLSILFIFGILSFYTGLLLRSCL---DSEPGLE--TY 215
I + G G+LS +A + GW +G S++ F ++++YT LL C D G T+
Sbjct: 785 ITAVVGSGVLSLAWAIAQLGWVIGPSVMIFFSLITWYTSSLLAECYRIGDPHYGKRNYTF 964
Query: 216 PDIGQAAFGTAGRIAISIVLYVELYGCCIEY 246
+ G G IV Y LYG I Y
Sbjct: 965 MEAGHTILGGFNDTLCGIVQYTNLYGTAIGY 1057
>BF631772 similar to PIR|F86385|F86 hypothetical protein AAG12739.1
[imported] - Arabidopsis thaliana, partial (29%)
Length = 523
Score = 45.1 bits (105), Expect = 7e-05
Identities = 34/120 (28%), Positives = 57/120 (47%), Gaps = 6/120 (5%)
Frame = +2
Query: 138 KKESKVSHEVPSRHCSFGQAVLNGINVLCGVGILSTPYAAKEGGW-LGLSILFIFGILSF 196
+ E K P+R + + + + + G G+LS PYA GW G+ +L + L+
Sbjct: 71 QSEQKEVENGPARRAKWWYSTFHTVTAMIGAGVLSLPYAMAYLGWGPGILMLLLSWCLTL 250
Query: 197 YTGLLLRSCLDSEPG--LETYPDIGQAAFGTAGRIAISIVLYVEL---YGCCIEYIILEG 251
T + + PG + Y D+G+ AFG ++ IVL +L GC I Y+++ G
Sbjct: 251 NTMWQMIQLHECVPGTRFDRYIDLGRHAFGP--KLGAWIVLPQQLIVQVGCDIVYMVIGG 424
>AL368579 similar to GP|14194151|gb AT5g40780/K1B16_3 {Arabidopsis thaliana},
partial (29%)
Length = 504
Score = 42.7 bits (99), Expect = 4e-04
Identities = 34/139 (24%), Positives = 66/139 (47%), Gaps = 9/139 (6%)
Frame = +1
Query: 125 TLLPPLSRRSSLLKKESKVSHEVP---SRHCSFGQAVLNGINVLCGVGILSTPYAAKEGG 181
T P + LK++ + +P SR+ + + + + + G G+LS PYA + G
Sbjct: 67 TQAPTATDIDEKLKRQKAIDDWLPISSSRNAKWWYSAFHNVTAMVGAGVLSLPYAMAQLG 246
Query: 182 W-LGLSILFIFGILSFYTGLLLRSCLDSEPG--LETYPDIGQAAFGTAGRIAISIVLYVE 238
W +++L + ++ YT + + PG + Y ++ Q AFG ++ + IV+ +
Sbjct: 247 WGPEVTVLVLSWFITLYTLWQMVEMHEMVPGKRFDRYHELSQYAFGE--KLGLYIVVPQQ 420
Query: 239 L---YGCCIEYIILEGDNL 254
L G I Y++ G +L
Sbjct: 421 LIVEVGVNIVYMVTGGKSL 477
>BF003758 similar to GP|15216030|em amino acid permease AAP4 {Vicia faba var.
minor}, partial (31%)
Length = 554
Score = 42.7 bits (99), Expect = 4e-04
Identities = 27/102 (26%), Positives = 48/102 (46%), Gaps = 6/102 (5%)
Frame = +2
Query: 162 INVLCGVGILSTPYAAKEGGWL-GLSILFIFGILSFYTGLLLRSCLDSEPGLE-----TY 215
I + G G+LS +A + GW+ G ++ +F +++YT +LL C + + TY
Sbjct: 233 ITAVIGSGVLSLAWAIAQLGWIAGPIVMILFAWVTYYTSILLCECYRTGDPISGKRNYTY 412
Query: 216 PDIGQAAFGTAGRIAISIVLYVELYGCCIEYIILEGDNLASL 257
D+ + G IV Y+ L G I Y I ++ ++
Sbjct: 413 MDVVHSNLGGFQVTLCGIVQYLNLVGVAIGYTIASAISMMAI 538
>TC93112 weakly similar to GP|6671946|gb|AAF23206.1| putative amino acid
transporter protein {Arabidopsis thaliana}, partial
(39%)
Length = 680
Score = 42.0 bits (97), Expect = 6e-04
Identities = 44/174 (25%), Positives = 73/174 (41%), Gaps = 23/174 (13%)
Frame = +3
Query: 359 SMAKPNQFPAVLVACFGVCTLLYAGGAVMGYKMFGEDTLSQFTLNLPQDLVATKIAVWTT 418
SM +FP +L F TL+Y + GY FGE+T TLNLP+ + + V
Sbjct: 39 SMKDKAKFPKLLAQTFSGITLVYILFGLCGYMAFGEETRDIVTLNLPRSWSSLSVQVGLC 218
Query: 419 VVNPFTKYALTISPVAMSLE---ELIPANHAKSY---LFSIFIRTGLVFSTL-VIGLSVP 471
+ FT + + + P+ +E ++I N+ S +++I +V L VI VP
Sbjct: 219 LGLMFT-FPIMLHPINEIVEGKLKIIRRNNNDSIRLGKITVYISRAIVVVVLAVIASFVP 395
Query: 472 FFVALCYGKLAITCLII----------------LCRFGDVLDWIITYNACNFDI 509
F + C ++ LC + VLD+I+ + F +
Sbjct: 396 EFGVFASFVGSTLCAMLSFVLPATFHLKLFGSSLCLWQKVLDYIVLISGLFFAV 557
>TC82248 similar to GP|15216028|emb|CAC51424. amino acid permease AAP3
{Vicia faba var. minor}, partial (29%)
Length = 779
Score = 38.1 bits (87), Expect = 0.009
Identities = 27/93 (29%), Positives = 41/93 (44%), Gaps = 6/93 (6%)
Frame = +3
Query: 162 INVLCGVGILSTPYAAKEGGWL-GLSILFIFGILSFYTGLLLRSCL---DSEPGLE--TY 215
I + G G+LS ++ + GW+ G + F I++ YT L C D+E G T+
Sbjct: 492 ITAVIGSGVLSLAWSIAQMGWVAGPGAMIFFSIITLYTSSFLADCYRCGDTEFGKRNYTF 671
Query: 216 PDIGQAAFGTAGRIAISIVLYVELYGCCIEYII 248
D G IV Y+ L+G + Y I
Sbjct: 672 MDAVSNILGGPSVKICGIVQYLNLFGSAMRYNI 770
>TC76989 homologue to SP|O24310|EFTU_PEA Elongation factor Tu chloroplast
precursor (EF-Tu). [Garden pea] {Pisum sativum}, partial
(92%)
Length = 1907
Score = 35.0 bits (79), Expect = 0.075
Identities = 33/124 (26%), Positives = 54/124 (42%), Gaps = 2/124 (1%)
Frame = +3
Query: 3 NSVASENSFIIESDEEDDKDFNKGDDGNDSDSSNYSNENPPQRKQSSYNPSWPQSYRQSI 62
NS S ++ ED+K N + + + S+ + + K SS P P ++ +
Sbjct: 54 NSSLISTSIHSKTKREDNKTNPLLFTLNSTMAISISSSSSTKLKLSSPYP--PHTHTFTT 227
Query: 63 DLYSSVPSPSIGFLGNSSLTRLSSSFLSTSLTRRHTPEVLPSVTK--PLIQPTEEEKHQR 120
+S+ +PS +LT LSSSFL+ S TP P+ T+ P K +R
Sbjct: 228 SSSASISNPSTTHRPKLTLTHLSSSFLNPSTILHLTPSSKPTPTRSSPFTVRAARGKFER 407
Query: 121 RSSH 124
+ H
Sbjct: 408 KKPH 419
>AW692849
Length = 658
Score = 33.9 bits (76), Expect = 0.17
Identities = 37/123 (30%), Positives = 52/123 (42%), Gaps = 12/123 (9%)
Frame = -2
Query: 36 NYSNENPPQRKQSSYNPSWPQSYRQSIDLYSSVPSPSIGFLG--NSSLTRL-SSSFLSTS 92
NYSN +N SW ++ S + SP L N S L S S LS
Sbjct: 456 NYSNY---------WNNSWQLNHNLSYESLILTVSPCFNLLAKPNPSFNILVSCSSLSLM 304
Query: 93 LTRRHTPEV----LPS---VTKPLIQPTE--EEKHQRRSSHTLLPPLSRRSSLLKKESKV 143
L +H + LPS + +P+++ E K + RSS L P+S ++ K K
Sbjct: 303 LQTQHKASIFTT*LPS*L*LLQPMLESLE*ISSKKKNRSSKLLKGPMSSKAQENSKACKK 124
Query: 144 SHE 146
SHE
Sbjct: 123 SHE 115
>AW586394 similar to GP|15216026|em amino acid permease AAP1 {Vicia faba var.
minor}, partial (42%)
Length = 613
Score = 32.7 bits (73), Expect = 0.37
Identities = 35/155 (22%), Positives = 53/155 (33%), Gaps = 20/155 (12%)
Frame = +1
Query: 339 AIGLYGYCYSGHAVFPNIYTSMAKP----NQFPAVLVACFGVCTLLYAGGAVMGYKMFGE 394
A+G + YS + I ++ P GV T Y MGY FG+
Sbjct: 70 ALGNIAFAYSYSQILIEIQDTIKNPPSEVKTMKQATKISIGVTTAFYMLCGCMGYAAFGD 249
Query: 395 DTLSQFTLNLPQDLVATKIAVWTTVVNPFTKYALTISPVAMSLEEL-------------- 440
+ IA V++ Y + P +E++
Sbjct: 250 TAPGNLLTGIFNPYWLIDIANAAIVIHLVGAYQVYAQPFFAFVEKIVIKRWPKINKEYRI 429
Query: 441 -IPANHAKSY-LFSIFIRTGLVFSTLVIGLSVPFF 473
IP H + LF + RT V +T VI + +PFF
Sbjct: 430 PIPGFHPYNLNLFRLIWRTIFVITTTVIAMLIPFF 534
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,097,430
Number of Sequences: 36976
Number of extensions: 334129
Number of successful extensions: 2171
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 2111
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2158
length of query: 552
length of database: 9,014,727
effective HSP length: 101
effective length of query: 451
effective length of database: 5,280,151
effective search space: 2381348101
effective search space used: 2381348101
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144720.2


