

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144657.4 + phase: 0
(776 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
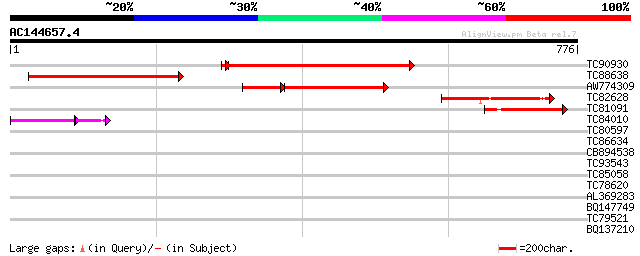
Score E
Sequences producing significant alignments: (bits) Value
TC90930 similar to GP|1742955|emb|CAA96058.1 CLC-b chloride chan... 296 1e-81
TC88638 similar to PIR|T02939|T02939 chloride channel protein Cl... 278 7e-75
AW774309 similar to GP|1742957|emb CLC-c chloride channel protei... 174 3e-55
TC82628 similar to GP|1742959|emb|CAA96065.1 CLC-d chloride chan... 118 1e-26
TC81091 similar to GP|4768916|gb|AAD29679.1| CLC-Nt2 protein {Ni... 109 5e-24
TC84010 similar to GP|1742959|emb|CAA96065.1 CLC-d chloride chan... 39 2e-04
TC80597 similar to GP|22136776|gb|AAM91732.1 unknown protein {Ar... 27 0.21
TC86634 similar to GP|9759532|dbj|BAB10998.1 outer membrane lipo... 30 2.7
CB894538 similar to GP|21553811|gb| outer membrane lipoprotein-l... 30 2.7
TC93543 weakly similar to SP|P48786|PRH_PETCR Pathogenesis-relat... 30 3.5
TC85058 similar to PIR|T47906|T47906 FUSCA PROTEIN FUS6 - Arabid... 30 4.6
TC78620 similar to PIR|T02336|T02336 cytochrome P450 homolog F13... 30 4.6
AL369283 homologue to GP|22136432|gb| putative protein {Arabidop... 29 7.8
BQ147749 similar to GP|18148451|db flavonoid 3'-hydroxylase {Gly... 29 7.8
TC79521 similar to GP|19310489|gb|AAL84978.1 AT4g39730/T19P19_12... 29 7.8
BQ137210 similar to SP|P09789|GRP1_ Glycine-rich cell wall struc... 29 7.8
>TC90930 similar to GP|1742955|emb|CAA96058.1 CLC-b chloride channel protein
{Arabidopsis thaliana}, partial (35%)
Length = 857
Score = 296 bits (758), Expect(2) = 1e-81
Identities = 136/257 (52%), Positives = 195/257 (74%), Gaps = 2/257 (0%)
Frame = +1
Query: 300 KGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICR 359
+ G IMFD + ++ YH++D+ PV ++ ++GGLLGSL+N + +KVLR+YN+IN+KG + +
Sbjct: 34 QAGXIMFDVSNVTVRYHVMDILPVAVIGIIGGLLGSLYNHLLHKVLRLYNLINQKGKMHK 213
Query: 360 LFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEP--CPTIGRSGIYKKFQCPPNHYNGLA 417
L L+ + +FTS +GLP+LA C PC P E CPT GRSG +K+F CPP HYN LA
Sbjct: 214 LLLSLAVVLFTSACQYGLPFLAKCTPCNPSLPESELCPTNGRSGNFKQFNCPPGHYNDLA 393
Query: 418 SLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGA 477
+L+ TNDDA+RN+FS +T +E++ SS+L+FF + L + + GI P GLF+PII+ G+
Sbjct: 394 TLLLTTNDDAVRNIFSSNTPHEYQTSSLLIFFALYCILGLITFGIAVPTGLFLPIILMGS 573
Query: 478 SYGRLVGILVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMV 537
+YGRLVG+L+G T + GL+AVLGAASL+ GSMR TVSLCVI LELT+NLLLLP+ M+V
Sbjct: 574 AYGRLVGMLMGPHTKIDQGLFAVLGAASLMAGSMRMTVSLCVIFLELTSNLLLLPITMIV 753
Query: 538 LVVSKSVANVFNANVYD 554
L+++K+V + FN ++Y+
Sbjct: 754 LLIAKTVGDCFNPSIYE 804
Score = 26.2 bits (56), Expect(2) = 1e-81
Identities = 10/13 (76%), Positives = 10/13 (76%)
Frame = +3
Query: 290 CLSDKCGLFGKGG 302
C S KCGLFG GG
Sbjct: 3 CNSGKCGLFGAGG 41
>TC88638 similar to PIR|T02939|T02939 chloride channel protein ClC-1 -
common tobacco, partial (29%)
Length = 1585
Score = 278 bits (710), Expect = 7e-75
Identities = 129/212 (60%), Positives = 163/212 (76%)
Frame = +3
Query: 27 INSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVVQILQYICMKWLLCFMIGLIV 86
IN+TSQ+AIVG+N+CPIESLDYE+F+NE F QDWRSR VQI QY+ +KW+ +IGL
Sbjct: 489 INTTSQIAIVGSNLCPIESLDYEVFDNEMFNQDWRSRKRVQIFQYVVLKWVFALLIGLGT 668
Query: 87 GFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFFASNLSLTLFASIITAFIAPTA 146
G +G NN++VEN+AG K T+++M ++RF AF + N+ L A+ + AFIAP+A
Sbjct: 669 GLVGLFNNISVENIAGFKLTLTTDLMSKQRFFEAFLAYAGLNMVLAAAAAALCAFIAPSA 848
Query: 147 AGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAALL 206
AGSGI EVKAYLNG+DA I TL VKI+GSI VS V+GK GPMVHTGAC+A++L
Sbjct: 849 AGSGIPEVKAYLNGIDAHSILAPTTLFVKIVGSILGVSAGFVVGKEGPMVHTGACIASIL 1028
Query: 207 GQGGSKRYGITWRWLRFFKNDRDRRDLIICGS 238
GQGGSK+YG+TW WLR+FKNDRDRRD+I C S
Sbjct: 1029GQGGSKKYGLTWSWLRYFKNDRDRRDMISCRS 1124
Score = 35.8 bits (81), Expect = 0.064
Identities = 12/22 (54%), Positives = 17/22 (76%)
Frame = +2
Query: 738 TRHDFTPEHILGMHPFLVKSRW 759
TRHDF PEH+LG++P + +W
Sbjct: 1100 TRHDFMPEHVLGLYPDIKHHKW 1165
>AW774309 similar to GP|1742957|emb CLC-c chloride channel protein
{Arabidopsis thaliana}, partial (22%)
Length = 658
Score = 174 bits (441), Expect(2) = 3e-55
Identities = 80/142 (56%), Positives = 101/142 (70%)
Frame = +1
Query: 377 LPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFSMHT 436
LPW+A C PCP D+ PCP++ SG YK FQCPP +YN LASL NTNDDAIRNLFS
Sbjct: 232 LPWIANCIPCPTDSKVPCPSVDESGEYKIFQCPPGYYNDLASLFLNTNDDAIRNLFSPKI 411
Query: 437 DNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNLSNG 496
EF +SS+ +FF FL I + GI P+GLF+P+I+ GA+YGR+V L T L G
Sbjct: 412 TKEFHISSLFIFFATVYFLGILTYGIAVPSGLFIPVILAGAAYGRVVSRLFEPITQLDRG 591
Query: 497 LYAVLGAASLLGGSMRTTVSLC 518
+++LGAAS+LGG+MR TVS+C
Sbjct: 592 FFSLLGAASMLGGTMRMTVSIC 657
Score = 60.1 bits (144), Expect(2) = 3e-55
Identities = 27/59 (45%), Positives = 40/59 (67%)
Frame = +3
Query: 319 DVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLLFGL 377
D+ V L V G+L S++NF+ +KV+R Y++INEKG ++FLA IS+ TSC + L
Sbjct: 57 DIVAVIGLGTVAGILASIYNFLVDKVVRTYSIINEKGPFFKIFLAVAISLLTSCCYYFL 233
>TC82628 similar to GP|1742959|emb|CAA96065.1 CLC-d chloride channel protein
{Arabidopsis thaliana}, partial (21%)
Length = 881
Score = 118 bits (295), Expect = 1e-26
Identities = 69/159 (43%), Positives = 97/159 (60%), Gaps = 5/159 (3%)
Frame = +1
Query: 592 IEKVRNIVFILRTTAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAF-----LPSP 646
+ KV ++V IL++ HNGFPVID E P++ G++LR HL +L+ K LPS
Sbjct: 58 VVKVSDVVSILQSNTHNGFPVIDHTRSGE-PLVIGLVLRSHLLVILQSKVDFQHSPLPSD 234
Query: 647 VANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSL 706
+R S +FAK S + + ++DI LT E++ M++DL PF N SPY V E MSL
Sbjct: 235 PRTGGRSIRH-DSGEFAKPVSSKGICLDDIHLTSEDLEMYIDLAPFLNPSPYIVPEDMSL 411
Query: 707 AKALILFREVGLRHLLVIPKIPGRSPVVGILTRHDFTPE 745
K LFR++GLRHL V+P+ P R V+G++TR D E
Sbjct: 412 TKVYNLFRQLGLRHLFVVPR-PSR--VLGLITRKDLLIE 519
>TC81091 similar to GP|4768916|gb|AAD29679.1| CLC-Nt2 protein {Nicotiana
tabacum}, partial (13%)
Length = 662
Score = 109 bits (272), Expect = 5e-24
Identities = 58/115 (50%), Positives = 82/115 (70%), Gaps = 2/115 (1%)
Frame = +3
Query: 651 YDVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKAL 710
++V KF+ + A++ KIE++ +T EEM MFVDLHP TN +P+TV+E++S+AKA+
Sbjct: 3 WEVREKFTWVELAEREG----KIEEVAITREEMEMFVDLHPLTNTTPFTVLESISVAKAM 170
Query: 711 ILFREVGLRHLLVIPK--IPGRSPVVGILTRHDFTPEHILGMHPFLVKSRWKRLR 763
ILFR+VGLRHLLV+PK G PV+GILTR D +IL + P L KS+ ++ R
Sbjct: 171 ILFRQVGLRHLLVVPKYQASGVCPVIGILTRQDLLAYNILTVFPHLAKSKNRQKR 335
>TC84010 similar to GP|1742959|emb|CAA96065.1 CLC-d chloride channel protein
{Arabidopsis thaliana}, partial (12%)
Length = 589
Score = 38.5 bits (88), Expect(2) = 2e-04
Identities = 29/99 (29%), Positives = 47/99 (47%), Gaps = 4/99 (4%)
Frame = +1
Query: 1 MSNNHLSNG-ESEPLLRRPLLSSQRS-IINSTSQVAIVGANV-CPIESLDYEIFENEFFK 57
M +NH+ NG ES L + +S S + V I+ N +ESLDYE+ EN ++
Sbjct: 163 MLSNHIQNGIESARLAWSRIPNSDESPFVLDDDGVGILKKNDGSAVESLDYEVIENFAYR 342
Query: 58 QDWRSRGVVQILQY-ICMKWLLCFMIGLIVGFIGFCNNL 95
++ R + Y + +KW +IG+ G N+
Sbjct: 343 EEQAHRRRKLYVSYLLVVKWFFALLIGICTGLAAVFINI 459
Score = 25.0 bits (53), Expect(2) = 2e-04
Identities = 15/47 (31%), Positives = 24/47 (50%)
Frame = +2
Query: 91 FCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFFASNLSLTLFASI 137
F + VEN AG K+ T N++ + + + I+F L +LF I
Sbjct: 446 FSSIFTVENFAGWKYSVTFNIIQKSYVRWLY*IYF-DQLGFSLFLRI 583
>TC80597 similar to GP|22136776|gb|AAM91732.1 unknown protein {Arabidopsis
thaliana}, partial (75%)
Length = 804
Score = 27.3 bits (59), Expect(2) = 0.21
Identities = 13/42 (30%), Positives = 27/42 (63%)
Frame = -2
Query: 512 RTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVY 553
R + +C++ L+L NLL+ L++M+L ++ VF+ +V+
Sbjct: 296 RQKIGICLVFLQLRVNLLVANLLLMLLYPTEV---VFSGSVF 180
Score = 25.4 bits (54), Expect(2) = 0.21
Identities = 11/24 (45%), Positives = 16/24 (65%)
Frame = -3
Query: 449 FIICLFLSIFSCGIVAPAGLFVPI 472
+I+ L +S+F C IV P F+PI
Sbjct: 403 YIL*L*ISVFECRIVCPEVKFLPI 332
>TC86634 similar to GP|9759532|dbj|BAB10998.1 outer membrane
lipoprotein-like {Arabidopsis thaliana}, partial (95%)
Length = 857
Score = 30.4 bits (67), Expect = 2.7
Identities = 16/36 (44%), Positives = 22/36 (60%), Gaps = 6/36 (16%)
Frame = +3
Query: 76 WLLCFMIGLIVGFIGFCN------NLAVENLAGIKF 105
+L CF+ L++G IGFC L ++NLAGI F
Sbjct: 312 FLQCFLSFLLLGIIGFCTLIMIIIMLLLDNLAGIIF 419
>CB894538 similar to GP|21553811|gb| outer membrane lipoprotein-like
{Arabidopsis thaliana}, partial (20%)
Length = 774
Score = 30.4 bits (67), Expect = 2.7
Identities = 16/36 (44%), Positives = 22/36 (60%), Gaps = 6/36 (16%)
Frame = +2
Query: 76 WLLCFMIGLIVGFIGFCN------NLAVENLAGIKF 105
+L CF+ L++G IGFC L ++NLAGI F
Sbjct: 2 FLQCFLSFLLLGIIGFCTLIMIIIMLLLDNLAGIIF 109
>TC93543 weakly similar to SP|P48786|PRH_PETCR Pathogenesis-related
homeodomain protein (PRHP). [Parsley Petroselinum
hortense], partial (4%)
Length = 903
Score = 30.0 bits (66), Expect = 3.5
Identities = 18/37 (48%), Positives = 23/37 (61%)
Frame = -1
Query: 288 DVCLSDKCGLFGKGGLIMFDAYSASISYHLVDVPPVF 324
DVC SDK LF + G+ FD +S S+ LVD P +F
Sbjct: 675 DVCGSDKRFLFFELGV--FDFFSTSLLGKLVDAPSLF 571
>TC85058 similar to PIR|T47906|T47906 FUSCA PROTEIN FUS6 - Arabidopsis
thaliana, partial (25%)
Length = 637
Score = 29.6 bits (65), Expect = 4.6
Identities = 11/28 (39%), Positives = 16/28 (56%)
Frame = +1
Query: 596 RNIVFILRTTAHNGFPVIDEPPGSEAPI 623
R I+ +R T H G P+ P G E+P+
Sbjct: 163 RQIISAVRPTRHRGIPLASTPAGPESPV 246
>TC78620 similar to PIR|T02336|T02336 cytochrome P450 homolog F13P17.32 -
Arabidopsis thaliana, partial (91%)
Length = 2058
Score = 29.6 bits (65), Expect = 4.6
Identities = 16/50 (32%), Positives = 28/50 (56%)
Frame = -2
Query: 619 SEAPILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSV 668
S A IL + L HH+ ++ K FL +A+ + V + +DF +++SV
Sbjct: 677 STAEIL--VFLAHHVDKIVFTKEFLTKRMAHKMESVGLYIGEDFMREFSV 534
>AL369283 homologue to GP|22136432|gb| putative protein {Arabidopsis
thaliana}, partial (4%)
Length = 460
Score = 28.9 bits (63), Expect = 7.8
Identities = 14/25 (56%), Positives = 16/25 (64%)
Frame = +2
Query: 355 GTICRLFLACLISIFTSCLLFGLPW 379
G+ CRLFL L S F + LLF L W
Sbjct: 344 GSCCRLFLLELRSWFLNLLLFALHW 418
>BQ147749 similar to GP|18148451|db flavonoid 3'-hydroxylase {Glycine max},
partial (38%)
Length = 702
Score = 28.9 bits (63), Expect = 7.8
Identities = 23/90 (25%), Positives = 41/90 (45%), Gaps = 12/90 (13%)
Frame = +3
Query: 673 IEDIQLTEEEMGMFVDLH----------PFTNASPYTVVETMSLAKALILFREVGL--RH 720
+E+I L + + + + LH T SP +V ++A A++++R + L +
Sbjct: 6 LEEITLAQSSLKIILHLHFPVYTRNTI*ATTGMSPLIIVFA-TIAAAILIYRLLNLITKP 182
Query: 721 LLVIPKIPGRSPVVGILTRHDFTPEHILGM 750
L +P P P+VG L P H L +
Sbjct: 183 SLPLPPGPSPWPIVGNLPHMGPVPHHALAV 272
>TC79521 similar to GP|19310489|gb|AAL84978.1 AT4g39730/T19P19_120
{Arabidopsis thaliana}, partial (72%)
Length = 1011
Score = 28.9 bits (63), Expect = 7.8
Identities = 14/54 (25%), Positives = 25/54 (45%), Gaps = 4/54 (7%)
Frame = +2
Query: 54 EFFKQDWRSRG----VVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGI 103
E F WR + V + +++C W + F+ + I C+ + EN+ GI
Sbjct: 809 ECF*DVWRRKKDSCFVFSVCKFMCYFWHIIFIYNMYGSLISLCDAMHXENMLGI 970
>BQ137210 similar to SP|P09789|GRP1_ Glycine-rich cell wall structural
protein 1 precursor. [Petunia] {Petunia hybrida},
partial (13%)
Length = 990
Score = 28.9 bits (63), Expect = 7.8
Identities = 16/50 (32%), Positives = 23/50 (46%), Gaps = 5/50 (10%)
Frame = -3
Query: 344 VLRIYNVINEKGTICRLFLACLISIFTSCLLFGL-----PWLAPCRPCPP 388
+LR +G +F CL+ +F C LFG+ +L P P PP
Sbjct: 184 ILRAVAWSGARGGAWVVFFGCLVLVFCLCFLFGVVVGGFVFLPPPPPSPP 35
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.328 0.143 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,201,837
Number of Sequences: 36976
Number of extensions: 415540
Number of successful extensions: 3311
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 2424
Number of HSP's successfully gapped in prelim test: 112
Number of HSP's that attempted gapping in prelim test: 820
Number of HSP's gapped (non-prelim): 2623
length of query: 776
length of database: 9,014,727
effective HSP length: 104
effective length of query: 672
effective length of database: 5,169,223
effective search space: 3473717856
effective search space used: 3473717856
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144657.4


