

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144656.4 + phase: 0
(472 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
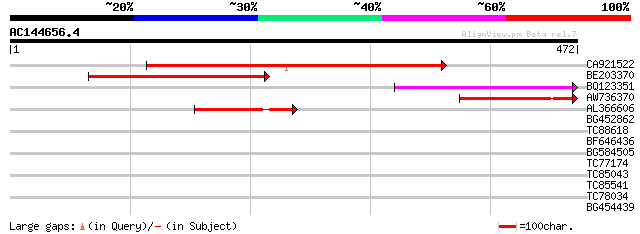
Score E
Sequences producing significant alignments: (bits) Value
CA921522 259 1e-69
BE203370 186 1e-47
BQ123351 94 1e-19
AW736370 77 2e-14
AL366606 77 2e-14
BG452862 weakly similar to GP|14715258|emb N3 like protein {Medi... 31 0.90
TC88618 weakly similar to PIR|T48397|T48397 S-receptor kinase-li... 29 4.5
BF646436 weakly similar to GP|9828634|gb F1N21.5 {Arabidopsis th... 29 4.5
BG584505 similar to GP|22831651|gb| CG32780-PA {Drosophila melan... 28 5.8
TC77174 similar to PIR|H84453|H84453 probable heat shock protein... 28 5.8
TC85043 28 7.6
TC85541 homologue to GP|166867|gb|AAA32866.1|| ribosomal protein... 28 9.9
TC78034 similar to GP|21592379|gb|AAM64330.1 unknown {Arabidopsi... 28 9.9
BG454439 similar to GP|14517399|gb At2g32810/F24L7.5 {Arabidopsi... 28 9.9
>CA921522
Length = 767
Score = 259 bits (663), Expect = 1e-69
Identities = 128/252 (50%), Positives = 170/252 (66%), Gaps = 3/252 (1%)
Frame = +3
Query: 115 EIADIKNKFITRAGLQCLPYNFLYQKATICFERSETDAFEAILALLIYGIVLFPNVDKFV 174
E ++K ITR +FLY++ T + AF +ILALL+YG+VLFP++D FV
Sbjct: 3 ETDEVKTHLITRGKFLGFSTDFLYERTTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFV 182
Query: 175 DMNAIQIFLTQNPVPTLLADTYVSIHERTDKERGTIVCCAPLLHIWITSHLPRPK---VK 231
D+ AIQIFL++NPVPTLL DTY+SIH RT RGTI+CCA LL+ WITSHLPR
Sbjct: 183 DIKAIQIFLSRNPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSHLPRTPRFTTN 362
Query: 232 PEYLPWSQKLMTLTPNDIVWFNPTCDPELIIDSCGDFNNVPLLGTRGGISYSPVLARRQF 291
PE L WS++LM+LTP ++VW++ D IIDSCG F NVPLLG GGISY+P LAR QF
Sbjct: 363 PENLLWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLARHQF 542
Query: 292 GFYMEMKPVYLILDRDFFLYKKDDANQRAQFKKAWYSIIRKDRNQLGNRSVIAHEAYVKW 351
G+ ME KP+ + L+ ++L D R +AW++I R+D++QLG ++ +Y +W
Sbjct: 543 GYPMERKPLSIYLENVYYLNADDSTGMREHVVRAWHTIRRRDKDQLGKKTGAISSSYTQW 722
Query: 352 VIDRANKWKMPY 363
VIDRA + MPY
Sbjct: 723 VIDRAVQIGMPY 758
>BE203370
Length = 454
Score = 186 bits (473), Expect = 1e-47
Identities = 95/151 (62%), Positives = 112/151 (73%)
Frame = +1
Query: 66 HCFTFSDYQLVPTLEEYSHWVGLPVLDQVPFHGLEPGPKIPAIANALHLEIADIKNKFIT 125
HCFTF DYQL PTLEEYS+ VG PVLD+VPF G EP PK IA+ALHLE + IK K
Sbjct: 1 HCFTFPDYQLAPTLEEYSYLVG*PVLDKVPFTGFEPIPKAATIADALHLETSLIKAKLTI 180
Query: 126 RAGLQCLPYNFLYQKATICFERSETDAFEAILALLIYGIVLFPNVDKFVDMNAIQIFLTQ 185
+ L LP FLYQ+A+ + DAF +ILALLIYG+VLFPNVD +VD++AIQ+FLT+
Sbjct: 181 KGNLPSLPTKFLYQQASDFAKVDNVDAFYSILALLIYGLVLFPNVDNYVDIHAIQMFLTK 360
Query: 186 NPVPTLLADTYVSIHERTDKERGTIVCCAPL 216
NPVPTLLAD Y SIH+R RG I+ CAPL
Sbjct: 361 NPVPTLLADMYHSIHDRNQVGRGAILGCAPL 453
>BQ123351
Length = 725
Score = 94.0 bits (232), Expect = 1e-19
Identities = 51/153 (33%), Positives = 86/153 (55%), Gaps = 1/153 (0%)
Frame = +2
Query: 321 QFKKAWYSIIRKDRNQLGNRSVIAHEAYVKWVIDRANKWKMPYPRQRLVTSTVSAIPLPL 380
+F +AW ++ ++++ QLG RS + +E+Y KWVIDRA K +P+P QRL +ST + LP+
Sbjct: 5 KFIQAWRAVHKREKGQLGRRS*LVYESYTKWVIDRAAKNGIPFPLQRLPSSTTLSPSLPM 184
Query: 381 PPESLEGYQKQLDIERRENSMWEVKYRKKKQEYDTVKNLLDQQIQANCKEKNENARLKAS 440
++ E Q L RE W ++Y + + E T++ ++Q+ K + +
Sbjct: 185 TSKTKEEAQYLLAEMTREKDTWRIRYMEAESEIGTLRGQVEQKDHELLKMRQQMIERDDL 364
Query: 441 IQRKEDFLDK-ICPGRKKRRMDLFNGPHLDSEE 472
+Q K+ L K I ++ MDLF+GP D E+
Sbjct: 365 LQEKDKLLKKHITKKQRMDSMDLFDGPDSDFED 463
>AW736370
Length = 488
Score = 76.6 bits (187), Expect = 2e-14
Identities = 42/98 (42%), Positives = 63/98 (63%)
Frame = -2
Query: 375 AIPLPLPPESLEGYQKQLDIERRENSMWEVKYRKKKQEYDTVKNLLDQQIQANCKEKNEN 434
A PLP+ ++ + YQ++L RE W+ +Y+KK Q+Y+TV LL+Q+ + +
Sbjct: 475 APPLPMTFDTEKEYQERLTEAEREIHRWKREYQKKDQDYETVMGLLEQEAYDSR*KDVII 296
Query: 435 ARLKASIQRKEDFLDKICPGRKKRRMDLFNGPHLDSEE 472
A+L I+ + LD+I PGRKKRRMDLF+GPH D E+
Sbjct: 295 AKLNERIKENDAALDRI-PGRKKRRMDLFDGPHSDFED 185
>AL366606
Length = 393
Score = 76.6 bits (187), Expect = 2e-14
Identities = 42/85 (49%), Positives = 56/85 (65%)
Frame = -1
Query: 155 AILALLIYGIVLFPNVDKFVDMNAIQIFLTQNPVPTLLADTYVSIHERTDKERGTIVCCA 214
AI L G++LFPN+D FVDMNAI++F ++NPVPTLLADTY +IH+RT K RG I+
Sbjct: 381 AIXLCLSTGLILFPNLDNFVDMNAIEVFHSKNPVPTLLADTYHAIHDRTLKGRGYIL--- 211
Query: 215 PLLHIWITSHLPRPKVKPEYLPWSQ 239
LHI ++ + + LP Q
Sbjct: 210 -WLHILFSNRVVYFRTFRSSLPMEQ 139
>BG452862 weakly similar to GP|14715258|emb N3 like protein {Medicago
truncatula}, partial (33%)
Length = 658
Score = 31.2 bits (69), Expect = 0.90
Identities = 21/87 (24%), Positives = 41/87 (46%)
Frame = +3
Query: 360 KMPYPRQRLVTSTVSAIPLPLPPESLEGYQKQLDIERRENSMWEVKYRKKKQEYDTVKNL 419
K+P + +V + + +P + E K+L++ + + E K K+ +E+D +
Sbjct: 372 KLPEHKGDIVDKEIENVVVPSKTTNDE---KKLEVSVVDMVIVEKKEEKQDEEHDEKEKK 542
Query: 420 LDQQIQANCKEKNENARLKASIQRKED 446
DQ Q K KNEN + + ++D
Sbjct: 543 QDQVTQDKTKVKNENDNININKTEEKD 623
>TC88618 weakly similar to PIR|T48397|T48397 S-receptor kinase-like protein
- Arabidopsis thaliana, partial (38%)
Length = 2304
Score = 28.9 bits (63), Expect = 4.5
Identities = 14/42 (33%), Positives = 23/42 (54%)
Frame = +2
Query: 184 TQNPVPTLLADTYVSIHERTDKERGTIVCCAPLLHIWITSHL 225
T N L DT++ ++E DKE+G+++ +L W S L
Sbjct: 452 TNNLYSLRLGDTFMGLYENHDKEKGSLLTKRLVLLYWKRSAL 577
>BF646436 weakly similar to GP|9828634|gb F1N21.5 {Arabidopsis thaliana},
partial (11%)
Length = 659
Score = 28.9 bits (63), Expect = 4.5
Identities = 15/49 (30%), Positives = 25/49 (50%)
Frame = +1
Query: 402 WEVKYRKKKQEYDTVKNLLDQQIQANCKEKNENARLKASIQRKEDFLDK 450
W+ K+ +KN+L Q+ + ++NE RLK Q ED+L +
Sbjct: 139 WDELDLKRADVEKELKNVLQQKEEILKLQQNEEERLKKEKQATEDYLQR 285
>BG584505 similar to GP|22831651|gb| CG32780-PA {Drosophila melanogaster},
partial (1%)
Length = 822
Score = 28.5 bits (62), Expect = 5.8
Identities = 17/62 (27%), Positives = 30/62 (47%)
Frame = +2
Query: 389 QKQLDIERRENSMWEVKYRKKKQEYDTVKNLLDQQIQANCKEKNENARLKASIQRKEDFL 448
+K+ + + N + ++++KKK E DT L QIQ E + +L+ KE
Sbjct: 122 EKEAIEKEKNNKLRPLQFQKKKIEDDTEAKLRPLQIQKQKIEDDAKVKLRKLQTEKEKIQ 301
Query: 449 DK 450
D+
Sbjct: 302 DE 307
>TC77174 similar to PIR|H84453|H84453 probable heat shock protein [imported] -
Arabidopsis thaliana, partial (90%)
Length = 2727
Score = 28.5 bits (62), Expect = 5.8
Identities = 18/63 (28%), Positives = 37/63 (58%), Gaps = 5/63 (7%)
Frame = +2
Query: 383 ESLEGYQKQ--LDIERRENSMW---EVKYRKKKQEYDTVKNLLDQQIQANCKEKNENARL 437
++L+ Y+++ +DI + + + EVK R+ KQEY+ + + + QQ+ N + + RL
Sbjct: 1865 QNLQTYKEKKFVDISKEDLELGDEDEVKERETKQEYNLLCDWIKQQLGDNVAKVQVSKRL 2044
Query: 438 KAS 440
+S
Sbjct: 2045 SSS 2053
>TC85043
Length = 308
Score = 28.1 bits (61), Expect = 7.6
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 2/64 (3%)
Frame = +3
Query: 266 GDFNNVPLLGTRGGISYSPVLARRQFGFY--MEMKPVYLILDRDFFLYKKDDANQRAQFK 323
GDF N G + + YSP+ RQ+G + + + +Y + FFL KK N +
Sbjct: 90 GDFCNYE--GKKKNVKYSPLELERQYGLFESIVLPNIYQVYPM-FFLIKKGYRNPAMKHY 260
Query: 324 KAWY 327
+Y
Sbjct: 261 SLYY 272
>TC85541 homologue to GP|166867|gb|AAA32866.1|| ribosomal protein S11
(probable start codon at bp 67) {Arabidopsis thaliana},
partial (87%)
Length = 883
Score = 27.7 bits (60), Expect = 9.9
Identities = 21/77 (27%), Positives = 37/77 (47%), Gaps = 2/77 (2%)
Frame = -1
Query: 89 PVLDQVPFHGLEPGPKIPAIAN--ALHLEIADIKNKFITRAGLQCLPYNFLYQKATICFE 146
P ++ +P G +P++A+ L+ D+KN+F GL LP F ++ CF
Sbjct: 316 PRMETLPVKGHFLSM*VPSMASLGVLNPRPTDLKNRFPPFPGLFPLPLFFEHRNTLGCFR 137
Query: 147 RSETDAFEAILALLIYG 163
+ AF A++ +G
Sbjct: 136 K----AFSVCSAMVKFG 98
>TC78034 similar to GP|21592379|gb|AAM64330.1 unknown {Arabidopsis
thaliana}, partial (73%)
Length = 947
Score = 27.7 bits (60), Expect = 9.9
Identities = 14/47 (29%), Positives = 24/47 (50%)
Frame = -2
Query: 15 KPDLESLRNLGRMVKNPEHFRNRYGYLLNILRTNVDEGLINTLVQFY 61
K ++ ++ L + + N H + ++ L TN+D GL NTL Y
Sbjct: 802 KDHMKWVKTLPKNILNNRHSMSLPEVIIQCLATNLDSGL*NTLETIY 662
>BG454439 similar to GP|14517399|gb At2g32810/F24L7.5 {Arabidopsis thaliana},
partial (29%)
Length = 669
Score = 27.7 bits (60), Expect = 9.9
Identities = 21/72 (29%), Positives = 32/72 (44%), Gaps = 13/72 (18%)
Frame = +3
Query: 217 LHIWITS---------HLPRPKVKPEYLPWSQKLMTLTPNDIVWFNPTCDPELIIDSCGD 267
++IWI S HL K + +YL +S ++ + + W P+L IDS D
Sbjct: 339 INIWINSSFTAEGIWEHLNVTKDQSDYLWYSTRIYVSDGDILFWKENAAHPKLAIDSVRD 518
Query: 268 ----FNNVPLLG 275
F N L+G
Sbjct: 519 ILRVFVNGQLIG 554
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.140 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,988,134
Number of Sequences: 36976
Number of extensions: 250163
Number of successful extensions: 1193
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 1185
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1190
length of query: 472
length of database: 9,014,727
effective HSP length: 100
effective length of query: 372
effective length of database: 5,317,127
effective search space: 1977971244
effective search space used: 1977971244
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144656.4


