

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144656.13 + phase: 0 /pseudo/partial
(1235 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
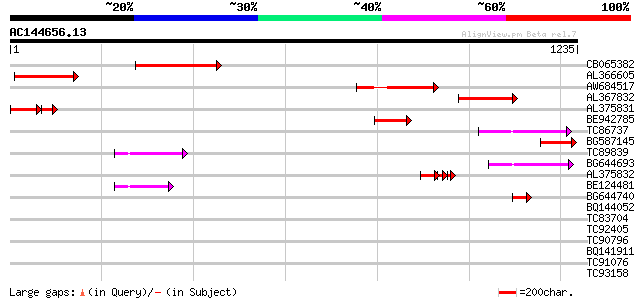
Score E
Sequences producing significant alignments: (bits) Value
CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fra... 318 8e-87
AL366605 273 2e-73
AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thalia... 251 1e-66
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 241 2e-63
AL375831 130 5e-46
BE942785 138 2e-32
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 92 1e-18
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 87 4e-17
TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC ... 84 3e-16
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 83 8e-16
AL375832 74 3e-13
BE124481 67 6e-11
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 44 4e-04
BQ144052 weakly similar to GP|21327989|dbj contains ESTs AU09387... 39 0.010
TC83704 similar to PIR|T05005|T05005 hypothetical protein T19P19... 37 0.036
TC92405 similar to GP|11994325|dbj|BAB02284. gene_id:MLM24.13~un... 37 0.047
TC90796 similar to GP|3395431|gb|AAC28763.1| unknown protein {Ar... 36 0.11
BQ141911 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 35 0.14
TC91076 similar to PIR|H96747|H96747 unknown protein T10D10.14 [... 35 0.18
TC93158 similar to PIR|T01123|T01123 hypothetical protein At2g32... 35 0.23
>CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fragment),
partial (9%)
Length = 624
Score = 318 bits (815), Expect = 8e-87
Identities = 155/189 (82%), Positives = 163/189 (86%), Gaps = 1/189 (0%)
Frame = -2
Query: 274 QPQVPVNAVVQQMQQQPPV-QQQQHQQARPTFPPIPMLYAELLPTLLLRGHCTTRQDKPP 332
+P +P PPV QQ+Q QQARPTFP IPMLYAE LPTLLLRGHCT RQ KPP
Sbjct: 569 RPMIP*LLC*PPRNNHPPVHQQRQ*QQARPTFPLIPMLYAE*LPTLLLRGHCTIRQGKPP 390
Query: 333 PDPLPPRFRSDLKCDFHQGALGHDVKGCYALKYIVKKLIDQGKLTFENNVPHVLDNPLPN 392
PDPLPPRFRSDLKCDFHQGALGHDV+GCYALK+IVKKLI+QGKLTFENNVPHVLDNPLPN
Sbjct: 389 PDPLPPRFRSDLKCDFHQGALGHDVEGCYALKHIVKKLINQGKLTFENNVPHVLDNPLPN 210
Query: 393 HAAVNMIEVYEEAPRLDVRNIATPLVPLHIKLCQASLFNHDHTNCPECFRNPLGCYVVQN 452
HAAVNMIEVYEEAP LDVRN+ TPLVPLHIKLCQASLF+HDH NC E F NPLGC VVQN
Sbjct: 209 HAAVNMIEVYEEAPGLDVRNVTTPLVPLHIKLCQASLFDHDHANCQE*FYNPLGCCVVQN 30
Query: 453 DIQSLMNGN 461
DIQSLMN N
Sbjct: 29 DIQSLMNNN 3
>AL366605
Length = 422
Score = 273 bits (699), Expect = 2e-73
Identities = 128/139 (92%), Positives = 133/139 (95%)
Frame = -3
Query: 11 EIKANRGNGDSVKTHDLCLVPKVDVPKKFKVPEFDRYNGLTCPHNHIIKYVRKMGNYSDN 70
EIKANRGN DS KT DLCLV KVDVPKKFK+P+FDRYNGLTCP NHIIKYVRKMGNY DN
Sbjct: 417 EIKANRGNADSFKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKDN 238
Query: 71 DSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQ 130
DSLMIHCFQDSLMEDAAEWYTSLSK+D+HTFDELAAAFKSHYGFNTRLKPNREFLRSLSQ
Sbjct: 237 DSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQ 58
Query: 131 KKDESFREYAQRWRGAAAR 149
KK+ESFREYAQRWRGAAAR
Sbjct: 57 KKEESFREYAQRWRGAAAR 1
>AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 488
Score = 251 bits (641), Expect = 1e-66
Identities = 131/180 (72%), Positives = 138/180 (75%)
Frame = +1
Query: 755 ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQNFKFVKNGKLVTVHGEEAYLVSQLSSF 814
ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ KFVKNG
Sbjct: 1 ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNG------------------- 123
Query: 815 SCIEAGSAEGTAFQGLTIEGMELKKTGTAMASLKDAQKAVQEGQAAGRGKLIQLRENKHK 874
SAEGTAFQGL++EG E KK G AMASLKDAQKAVQEGQAA GKLIQL ENK K
Sbjct: 124 ------SAEGTAFQGLSMEGAEPKKVGAAMASLKDAQKAVQEGQAADWGKLIQLCENKRK 285
Query: 875 EGLGFSPTSGVSTGTFHSAGFVNAITEEATGFGPRPVFVIPGGIARDWDAIDIPSIMHVS 934
EGL FSPTSGVSTGTFHSAGFVN + EE F PRP+FVIPGGIA+DWDA+D+PSIMHVS
Sbjct: 286 EGLRFSPTSGVSTGTFHSAGFVNTLAEEVARFVPRPLFVIPGGIAKDWDAVDVPSIMHVS 465
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 241 bits (614), Expect = 2e-63
Identities = 118/128 (92%), Positives = 122/128 (95%)
Frame = -1
Query: 978 QEKKAIQPHQEEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIFAWSYEDMP 1037
QE+KAIQPHQEEIE IN+GTEENKREIKVGAALEE VK+KI QLLREY DIFA SYEDMP
Sbjct: 384 QERKAIQPHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLLREYLDIFACSYEDMP 205
Query: 1038 GLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANI 1097
GLDPKIVEHRIPTKPECPPVR KLRRTHPDMALKIK+EVQKQIDAGFLMTVEYPEWVANI
Sbjct: 204 GLDPKIVEHRIPTKPECPPVR*KLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANI 25
Query: 1098 VPVPKKDG 1105
VPVPKKDG
Sbjct: 24 VPVPKKDG 1
>AL375831
Length = 467
Score = 130 bits (327), Expect(2) = 5e-46
Identities = 60/69 (86%), Positives = 63/69 (90%)
Frame = +2
Query: 1 MEELAKELRREIKANRGNGDSVKTHDLCLVPKVDVPKKFKVPEFDRYNGLTCPHNHIIKY 60
MEELAKELR EI+ANRGN DSVKT DLCLV KVDVPKKFK+P+FDRYNGLTCP NHIIKY
Sbjct: 158 MEELAKELRHEIQANRGNADSVKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKY 337
Query: 61 VRKMGNYSD 69
VRKMGNY D
Sbjct: 338 VRKMGNYKD 364
Score = 73.9 bits (180), Expect(2) = 5e-46
Identities = 32/34 (94%), Positives = 34/34 (99%)
Frame = +3
Query: 70 NDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDE 103
NDSLMIHCFQDSLMEDAAEWYTSLSK+D+HTFDE
Sbjct: 366 NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDE 467
>BE942785
Length = 460
Score = 138 bits (347), Expect = 2e-32
Identities = 68/82 (82%), Positives = 76/82 (91%)
Frame = -2
Query: 794 NGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGTAFQGLTIEGMELKKTGTAMASLKDAQKA 853
+G LVT+HGEEAYL+SQLSSFSCIEAGSAEGTAFQGLT+EG E K+ GTAMASLKDAQ+A
Sbjct: 378 SGXLVTIHGEEAYLISQLSSFSCIEAGSAEGTAFQGLTVEGTEPKRDGTAMASLKDAQRA 199
Query: 854 VQEGQAAGRGKLIQLRENKHKE 875
VQE QAAG G+LIQLRENKHK+
Sbjct: 198 VQESQAAGWGRLIQLRENKHKD 133
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 92.0 bits (227), Expect = 1e-18
Identities = 63/213 (29%), Positives = 105/213 (48%), Gaps = 9/213 (4%)
Frame = +1
Query: 1021 LLREYPDIFAWSYEDMPGLDPKIVEHRIPTKPEC----PPVRQ-KLRRTHPDMALKIKNE 1075
+L E+PD+F +++H IP P+ PP+ L L +K
Sbjct: 343 VLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKT 522
Query: 1076 VQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVD 1135
++ +D GF+ A ++ V K G +R CVD+R LN + KD +PL I +
Sbjct: 523 LEDLLDKGFIKASGSAAG-APVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLR 699
Query: 1136 NNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMT 1195
A ++ F +D + ++++++ ED+EKT+F T +G F + V P GL A AT+QR +
Sbjct: 700 RVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYIN 879
Query: 1196 TLFHDMIHKEVEVYVDDMIV----KSKDEEQHV 1224
H+ + V Y+DD+++ KD E V
Sbjct: 880 KTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQV 978
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 87.0 bits (214), Expect = 4e-17
Identities = 41/78 (52%), Positives = 55/78 (69%)
Frame = +2
Query: 1157 MSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVK 1216
M P+D EKT+FIT GT+CYKVMP GL NAG+TYQR + +F D + +EVY+DDM+VK
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 1217 SKDEEQHVEYLTKMFERL 1234
S H+ +L + F+ L
Sbjct: 182 SLRATDHLNHLKE*FKTL 235
>TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC 4.6.1.1)
(ATP pyrophosphate-lyase) (Adenylyl cyclase). [Yeast],
partial (0%)
Length = 665
Score = 84.0 bits (206), Expect = 3e-16
Identities = 57/159 (35%), Positives = 79/159 (48%)
Frame = +3
Query: 228 EVGMVSAGAGQSMATVAPINAA*LPPSYPYAPYSQHPFFPYPLPSGQPQVPVNAVVQQMQ 287
EV + A + + V+ + A PP P Q P+ P + Q P ++ Q
Sbjct: 156 EVTTLRAEVEKLTSLVSSLMATNDPPLVQQRP--QSPYQPMCPQKPRQQAPRQSIPQNQV 329
Query: 288 QQPPVQQQQHQQARPTFPPIPMLYAELLPTLLLRGHCTTRQDKPPPDPLPPRFRSDLKCD 347
Q + Q Q Q+A PIP+ YA+LLP LL + T P+ LPP +R DL C
Sbjct: 330 PQKFIPQNQVQKASQC-DPIPVKYADLLPILLKKNLIQTLPLPRVPNSLPPWYRPDLNCV 506
Query: 348 FHQGALGHDVKGCYALKYIVKKLIDQGKLTFENNVPHVL 386
FHQGA GHD + CY LK V+KLI+ +F++ VL
Sbjct: 507 FHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDDQDIKVL 623
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 82.8 bits (203), Expect = 8e-16
Identities = 56/184 (30%), Positives = 93/184 (50%)
Frame = +2
Query: 1044 VEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKK 1103
++ I P P+ R +P +K +++ ++ GF+ YP V ++ + KK
Sbjct: 35 IDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVV-VLFLKKK 211
Query: 1104 DGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDRE 1163
DG +RM +D+ LN + K +PL ID L DN SK FF +D G +Q ++ ED
Sbjct: 212 DGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGEDVP 391
Query: 1164 KTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQH 1223
KT+F +G + VM G N + M +F D + V V+ +D+++ SK+E +H
Sbjct: 392 KTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNENEH 571
Query: 1224 VEYL 1227
+L
Sbjct: 572 ENHL 583
>AL375832
Length = 535
Score = 73.9 bits (180), Expect = 3e-13
Identities = 35/41 (85%), Positives = 36/41 (87%)
Frame = +3
Query: 895 FVNAITEEATGFGPRPVFVIPGGIARDWDAIDIPSIMHVSE 935
FVNAI+EEATG G RP FV PGGIA DWDAIDIPSIMHVSE
Sbjct: 3 FVNAISEEATGSGLRPAFVTPGGIASDWDAIDIPSIMHVSE 125
Score = 32.0 bits (71), Expect(2) = 3e-04
Identities = 12/17 (70%), Positives = 14/17 (81%)
Frame = +3
Query: 955 FPMYEAEAEEGDDIPYE 971
FP+YE + EGDDIPYE
Sbjct: 483 FPVYEPKMREGDDIPYE 533
Score = 31.6 bits (70), Expect(2) = 3e-04
Identities = 13/22 (59%), Positives = 15/22 (68%)
Frame = +1
Query: 933 VSELNHNKPVGHSNPTFPPNFE 954
+ LNHNKPV HSN PNF+
Sbjct: 418 ICRLNHNKPVEHSNSMPLPNFD 483
>BE124481
Length = 538
Score = 66.6 bits (161), Expect = 6e-11
Identities = 46/129 (35%), Positives = 62/129 (47%)
Frame = +3
Query: 228 EVGMVSAGAGQSMATVAPINAA*LPPSYPYAPYSQHPFFPYPLPSGQPQVPVNAVVQQMQ 287
EV + A + + V+ + A PP P Q P+ P + Q P ++ Q
Sbjct: 159 EVTTLRAEVEKLTSLVSSLMATNDPPLVQQRP--QSPYQPMCPQKPRQQAPRQSIPQNQV 332
Query: 288 QQPPVQQQQHQQARPTFPPIPMLYAELLPTLLLRGHCTTRQDKPPPDPLPPRFRSDLKCD 347
Q + Q Q Q+A PIP+ YA+LLP LL + T P+ LPP +R DL C
Sbjct: 333 PQKFIPQNQVQKASQC-DPIPVKYADLLPILLKKNLIQTLPLPRVPNSLPPWYRPDLNCV 509
Query: 348 FHQGALGHD 356
FHQGA GHD
Sbjct: 510 FHQGAPGHD 536
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 43.9 bits (102), Expect = 4e-04
Identities = 20/41 (48%), Positives = 27/41 (65%)
Frame = -1
Query: 1095 ANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVD 1135
A ++ V KKDG RMC+D+R NK + K+ +PL ID L D
Sbjct: 238 AALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFD 116
>BQ144052 weakly similar to GP|21327989|dbj contains ESTs AU093876(E1018)
AU093877(E1018)~unknown protein, partial (11%)
Length = 536
Score = 39.3 bits (90), Expect = 0.010
Identities = 28/98 (28%), Positives = 39/98 (39%)
Frame = +1
Query: 252 PPSYPYAPYSQHPFFPYPLPSGQPQVPVNAVVQQMQQQPPVQQQQHQQARPTFPPIPMLY 311
PP +P P+ HP P+P P P +P PP+ P PP P
Sbjct: 265 PPQFP-PPFLPHPPLPFPFPIAPPPLPAIPC-------PPLIPA*IPSILPVSPPRPPPL 420
Query: 312 AELLPTLLLRGHCTTRQDKPPPDPLPPRFRSDLKCDFH 349
A + P L+L R +P P PR+ S ++ H
Sbjct: 421 A*IPPYLILTSPIALRLSQPRP---LPRYYSAIRITLH 525
>TC83704 similar to PIR|T05005|T05005 hypothetical protein T19P19.70 -
Arabidopsis thaliana, partial (15%)
Length = 716
Score = 37.4 bits (85), Expect = 0.036
Identities = 31/108 (28%), Positives = 43/108 (39%), Gaps = 9/108 (8%)
Frame = +1
Query: 241 ATVAPINAA*LPPSYPYAPYSQHPFFPYPLPSGQPQVPVNAVVQQMQQQPPVQQQQHQQA 300
+TV P+ LP PY + + P P LP P + ++ PP + A
Sbjct: 262 STVPPVATTSLPEPLPYQ-HKEQPPIPVTLPPPPPLSKLPPATREQLPPPPPLSKLPPAA 438
Query: 301 RPTFPPIPMLYAEL---------LPTLLLRGHCTTRQDKPPPDPLPPR 339
R PP P A+L LP L + R+ PPP PLP +
Sbjct: 439 RE-HPPPPPPSAKLPPAARERLPLPPPLSKLPPAARERLPPPSPLPEK 579
>TC92405 similar to GP|11994325|dbj|BAB02284. gene_id:MLM24.13~unknown
protein {Arabidopsis thaliana}, partial (69%)
Length = 864
Score = 37.0 bits (84), Expect = 0.047
Identities = 32/111 (28%), Positives = 49/111 (43%), Gaps = 1/111 (0%)
Frame = +2
Query: 688 KSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRISTFLVKAFDGSRKNV-LGEIDLPM 746
K+D + L D G +++ LD+L R + S F + GSR +G + LP+
Sbjct: 278 KADAAAKELEDAGGLVDLT--DNLDRLQGRWKLIYSSAFSSRTLGGSRPGPPIGRL-LPI 448
Query: 747 TIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQNFKFVKNGKL 797
T+G I D + LG PW VT+TL F+ V + K+
Sbjct: 449 TLGQVFQRIDILSKDFDNIVDLQLGAPWPLPPLEVTATLAHKFELVGSSKI 601
>TC90796 similar to GP|3395431|gb|AAC28763.1| unknown protein {Arabidopsis
thaliana}, partial (20%)
Length = 965
Score = 35.8 bits (81), Expect = 0.11
Identities = 24/64 (37%), Positives = 27/64 (41%), Gaps = 11/64 (17%)
Frame = +2
Query: 254 SYPYAPYS---QHPFFPYPLPSGQPQVPVNAVVQQMQQQ--------PPVQQQQHQQARP 302
SY YAP + P+ Y P QPQ QQ QQ PP QQ Q +P
Sbjct: 773 SYSYAPQAYPGNSPYNSYVAPWAQPQSKSEFQTQQPQQHMYQPQPTTPPSPQQHMYQPQP 952
Query: 303 TFPP 306
T PP
Sbjct: 953 TTPP 964
>BQ141911 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (16%)
Length = 1151
Score = 35.4 bits (80), Expect = 0.14
Identities = 24/72 (33%), Positives = 32/72 (44%), Gaps = 13/72 (18%)
Frame = -2
Query: 252 PPSYP---YAPYSQHPFF--PYPLPSGQPQVPVNAVVQQM--------QQQPPVQQQQHQ 298
PPS+P Y P +H F P+ SG P P N QQ Q PP+ + +
Sbjct: 289 PPSHPPTTYTPPHKHMFKTQPHVFRSGSPTQPPNQT*QQPTKHPLPQNQITPPLSKHDSK 110
Query: 299 QARPTFPPIPML 310
+ P PP+P L
Sbjct: 109 TSTPNSPPLPPL 74
>TC91076 similar to PIR|H96747|H96747 unknown protein T10D10.14 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 834
Score = 35.0 bits (79), Expect = 0.18
Identities = 29/100 (29%), Positives = 41/100 (41%), Gaps = 7/100 (7%)
Frame = +3
Query: 212 NASKRYGNGHHKKKETEVGMVSAGAGQSMATVAPI--NAA*LPPSYPYAPYSQHPFFPYP 269
N ++YG + E ++ G+ S+ + NA LPP P
Sbjct: 270 NEMQQYGEPIPGQSNNEAAKLAGGSNASLNLPQSLVANARMLPPGNPQGLQMSQALLSGV 449
Query: 270 LPSGQPQV--PVNAVVQQMQQQPPVQQQQH---QQARPTF 304
+ +PQ AV+QQ QQQ +QQ QH QQ P F
Sbjct: 450 SMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQF 569
>TC93158 similar to PIR|T01123|T01123 hypothetical protein At2g32840
[imported] - Arabidopsis thaliana, partial (9%)
Length = 673
Score = 34.7 bits (78), Expect = 0.23
Identities = 29/96 (30%), Positives = 34/96 (35%), Gaps = 2/96 (2%)
Frame = +2
Query: 245 PINAA*LPPSYPYAPYSQHPFFPYPLPSGQPQVPVNAVVQQMQQQPPVQQQQHQQARPTF 304
P A S P AP++ P P P+ +P P QQQ QQA
Sbjct: 44 PTTATTTTTSRPTAPFTN----PNPNPNSKPI-----------HYPQQQQQPQQQALHVR 178
Query: 305 PPIPMLYAELLPTLLLRGHCTTRQDKPPP--DPLPP 338
P P LY P+ H PPP P PP
Sbjct: 179 SPNPFLYPFASPSRASANHAVGGYPPPPPPSQPQPP 286
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,689,450
Number of Sequences: 36976
Number of extensions: 572396
Number of successful extensions: 3602
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 3322
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3526
length of query: 1235
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1128
effective length of database: 5,058,295
effective search space: 5705756760
effective search space used: 5705756760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144656.13


