

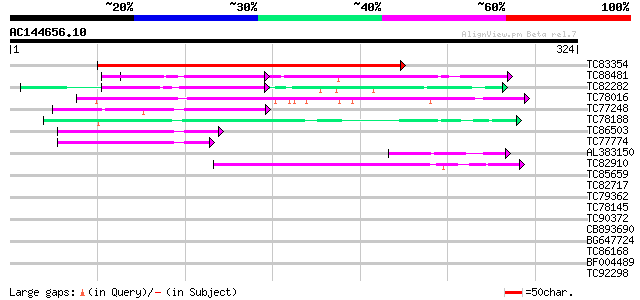
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144656.10 - phase: 0
(324 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83354 similar to GP|6227001|gb|AAF06037.1| Contains 3 PF|00400... 347 5e-96
TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclea... 57 7e-09
TC82282 similar to PIR|T08544|T08544 hypothetical protein F27B13... 48 4e-06
TC78016 similar to GP|21537191|gb|AAM61532.1 PRL1 protein {Arabi... 46 2e-05
TC77248 similar to GP|13489182|gb|AAK27816.1 putative WD-repeat ... 46 2e-05
TC78188 similar to PIR|C84870|C84870 probable splicing factor [i... 44 1e-04
TC86503 similar to GP|15028127|gb|AAK76687.1 unknown protein {Ar... 42 2e-04
TC77774 similar to GP|15028127|gb|AAK76687.1 unknown protein {Ar... 42 3e-04
AL383150 similar to GP|17529236|gb putative SecA-type chloroplas... 42 3e-04
TC82910 similar to GP|19347844|gb|AAL86002.1 unknown protein {Ar... 42 3e-04
TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein ... 40 0.001
TC82717 weakly similar to GP|21539583|gb|AAM53344.1 putative coa... 40 0.002
TC79362 WD-repeat cell cycle regulatory protein 39 0.002
TC78145 similar to GP|15294240|gb|AAK95297.1 AT3g18860/MCB22_3 {... 39 0.003
TC90372 similar to GP|20197166|gb|AAC27402.2 expressed protein {... 39 0.003
CB893690 weakly similar to GP|9759025|dbj gene_id:K6A12.9~simila... 37 0.004
BG647724 similar to GP|11141605|gb LEUNIG {Arabidopsis thaliana}... 37 0.010
TC86168 homologue to GP|13936812|gb|AAK49947.1 TGF-beta receptor... 37 0.013
BF004489 similar to GP|20161344|d contains ESTs AU086067(S4642) ... 37 0.013
TC92298 similar to PIR|T00642|T00642 hypothetical protein F3I6.5... 36 0.023
>TC83354 similar to GP|6227001|gb|AAF06037.1| Contains 3 PF|00400 WD40
G-beta repeat domains. {Arabidopsis thaliana}, partial
(41%)
Length = 531
Score = 347 bits (889), Expect = 5e-96
Identities = 174/176 (98%), Positives = 175/176 (98%)
Frame = +3
Query: 51 SVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLA 110
SV ASGGSDDTIHLYDLS+SSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLA
Sbjct: 3 SVGASGGSDDTIHLYDLSSSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLA 182
Query: 111 IFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKE 170
IFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKE
Sbjct: 183 IFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKE 362
Query: 171 ASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVLCAAPAKNGLLYTGG 226
ASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVLCAAPAKNGLLYTGG
Sbjct: 363 ASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVLCAAPAKNGLLYTGG 530
>TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclear
ribonucleoprotein [imported] - Arabidopsis thaliana,
partial (60%)
Length = 1836
Score = 57.4 bits (137), Expect = 7e-09
Identities = 60/229 (26%), Positives = 97/229 (42%), Gaps = 5/229 (2%)
Frame = +1
Query: 64 LYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTL 123
L+ + + +L H+ T ++ YSP + +L +A AD + ++ G + L T
Sbjct: 1027 LWSMPNVKKVSTLKGHTQRATDVA-YSPVH---KNHLATASADRTAKYWNDQGAL-LGTF 1191
Query: 124 SVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKEASIVRFDVSGDSF 183
H + + +A HPSGK T S D + + + V + S+ D D
Sbjct: 1192 KGHLERLARIAFHPSGKYLGTASYDKTWRLWD-VETEEELLLQEGHSRSVYGLDFHHDGS 1368
Query: 184 FMA---VDEIVNVHQAEDARLLMELQCP-KRVLCAAPAKNGL-LYTGGEDRNITAWDLKS 238
A +D + V R ++ L+ K +L + + NG L TGGED WDL+
Sbjct: 1369 LAASCGLDALARVWDLRTGRSVLALEGHVKSILGISFSPNGYHLATGGEDNTCRIWDLRK 1548
Query: 239 GKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAWDVR 287
K Y I AH+ ++S E Y + +AS D T + W R
Sbjct: 1549 KKSLYTI-PAHSN------LISQVKFEPQEGYFLVTASYDMTAKVWSSR 1674
Score = 47.8 bits (112), Expect = 6e-06
Identities = 31/96 (32%), Positives = 51/96 (52%)
Frame = +1
Query: 53 VASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIF 112
+A+GG D+T ++DL SL ++ HS+ ++ + F P F LV+A D + ++
Sbjct: 1498 LATGGEDNTCRIWDLRKKKSLYTIPAHSNLISQVKF-EPQEGYF---LVTASYDMTAKVW 1665
Query: 113 DADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRD 148
+ F +KTLS H+ V L + G +TVS D
Sbjct: 1666 SSRDFKPVKTLSGHEAKVTSLDVLGDGGYIVTVSHD 1773
>TC82282 similar to PIR|T08544|T08544 hypothetical protein F27B13.70 -
Arabidopsis thaliana, partial (84%)
Length = 904
Score = 48.1 bits (113), Expect = 4e-06
Identities = 33/96 (34%), Positives = 49/96 (50%)
Frame = +2
Query: 53 VASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIF 112
+A G D TI ++D+ + L L H V +L YSP + PR L SA DG++ ++
Sbjct: 593 LACGSMDGTISVFDVQRAKFLHHLEGHFMPVRSL-VYSPYD---PRLLFSASDDGNVHMY 760
Query: 113 DADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRD 148
DA+G T+S H V + + P+G T S D
Sbjct: 761 DAEGKALXGTMSXHASWVLCVDVSPNGAAIATGSSD 868
Score = 43.1 bits (100), Expect = 1e-04
Identities = 67/302 (22%), Positives = 109/302 (35%), Gaps = 24/302 (7%)
Frame = +2
Query: 7 ISLVAGSYERFIWGFNLNPNTQTLTPVFSYPSHLSLIKTVAVSNSVVASGGSDDTIHLYD 66
I + +++ +W P T T P+ + +G D+T+ L+
Sbjct: 92 IKSIDNAHDDSVWAVTWAPATATRPPL-------------------LLTGSLDETVRLW- 211
Query: 67 LSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVH 126
S + + ++T L S P S+ D + +FD D + TL
Sbjct: 212 ----KSDDLVLERTNTGHCLGVASVAAHPLGSIAASSSLDSFVRVFDVDSNATIATLEAP 379
Query: 127 KKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKEASIVRF------DVSG 180
V + P G I S A VNL + L SI R D SG
Sbjct: 380 PSEVWQMRFDPKGAILAVAGGGS--ASVNLWD---TSTWELVVTLSIPRVEGPKPSDKSG 544
Query: 181 DSFFM---------------AVDEIVNVHQAEDARLLMELQ---CPKRVLCAAPAKNGLL 222
F+ ++D ++V + A+ L L+ P R L +P LL
Sbjct: 545 SKKFVLSVAWSPDGKRLACGSMDGTISVFDVQRAKFLHHLEGHFMPVRSLVYSPYDPRLL 724
Query: 223 YTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIR 282
++ +D N+ +D + GK HA+ V + V + A +A+ SSD T+R
Sbjct: 725 FSASDDGNVHMYDAE-GKALXGTMSXHASWVLCVDVSPNGAA-------IATGSSDKTVR 880
Query: 283 AW 284
W
Sbjct: 881 LW 886
>TC78016 similar to GP|21537191|gb|AAM61532.1 PRL1 protein {Arabidopsis
thaliana}, partial (81%)
Length = 1759
Score = 46.2 bits (108), Expect = 2e-05
Identities = 72/311 (23%), Positives = 126/311 (40%), Gaps = 52/311 (16%)
Frame = +1
Query: 39 HLSLIKTVAV--SNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPF 96
HL +++VAV SN+ A+G +D TI ++DL++ +LT H V L+ F
Sbjct: 583 HLGWVRSVAVDPSNTWFATGSADRTIKIWDLASGVLKLTLTGHIEQVRGLAISHKHTYMF 762
Query: 97 PRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSC------ 150
SA D + +D + +++ H GV LAIHP+ I LT C
Sbjct: 763 -----SAGDDKQVKCWDLEQNKVIRSYHGHLSGVYCLAIHPTIDILLTGRT*FCLPGSGT 927
Query: 151 ------FAMVNLVR--------GRR------------SFCCRLD--KEASIVRFDVSGDS 182
F + ++R G+R S C LD K+ +++ S
Sbjct: 928 YAVKCRFMLFQVMRTLSALCLHGQRTLKLSLVLTILQSRCGTLDMVKQCQLLQTIKSLFE 1107
Query: 183 FFMAV--DEIVNVH----------QAEDARLLMELQCPKRVLCAAPAKNGLLYTGGEDRN 230
++++ +++++H Q E+ + L + + L GG++ +
Sbjct: 1108QWLSILKSKLLHLHQLIILKSSPFQKENFVTICSLNRKLLSMQWLSMRRVLWLPGGDNGS 1287
Query: 231 ITAWDLKSG----KVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAWDV 286
+ WD KSG + ++ GI L+ + TG + S +D TI+ W
Sbjct: 1288MWFWDWKSGHNFQQSQTIVQPGSLDSEAGIYALTYDVTGT----RLISCEADKTIKMWKE 1455
Query: 287 RMAATEKSEPL 297
AT ++ PL
Sbjct: 1456DDNATPETHPL 1488
>TC77248 similar to GP|13489182|gb|AAK27816.1 putative WD-repeat containing
protein {Oryza sativa}, partial (92%)
Length = 2005
Score = 46.2 bits (108), Expect = 2e-05
Identities = 34/128 (26%), Positives = 57/128 (43%), Gaps = 3/128 (2%)
Frame = +1
Query: 25 PNTQTLTPVFSYPSHLSLIKTVAVSNSVVASGGSDDTIHLYDLSTSSSLGS---LTDHSS 81
P+ Q L + + ++ +K V S++ +G +D T+ L+ S L DHS+
Sbjct: 850 PSGQVLATLTGHSKKVTSVKFVGQGESII-TGSADKTVRLWQGSDDGHYNCKQILKDHSA 1026
Query: 82 TVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKI 141
V A++ ++ N V+A DGS ++ L +S +G A HP G I
Sbjct: 1027EVEAVTVHATNNY-----FVTASLDGSWCFYELSSGTCLTQVSDSSEGYTSAAFHPDGLI 1191
Query: 142 ALTVSRDS 149
T + DS
Sbjct: 1192LGTGTTDS 1215
>TC78188 similar to PIR|C84870|C84870 probable splicing factor [imported] -
Arabidopsis thaliana, partial (95%)
Length = 1436
Score = 43.5 bits (101), Expect = 1e-04
Identities = 63/277 (22%), Positives = 104/277 (36%), Gaps = 4/277 (1%)
Frame = +1
Query: 20 GFNLNPNTQTL-TPVFSYPSHLSLIKTVAVS--NSVVASGGSDDTIHLYDL-STSSSLGS 75
G N T +L +P+ H S + T+ + SVVASG D I L+++ +
Sbjct: 250 GPNGKQRTSSLESPIMLLTGHQSAVYTMKFNPTGSVVASGSHDKEIFLWNVHGDCKNFMV 429
Query: 76 LTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAI 135
L H + V L + S ++SA D +L ++D + +K + H VN
Sbjct: 430 LKGHKNAVLDLHWTSDGT-----QIISASPDKTLRLWDTETGKQIKKMVEHLSYVNSCCP 594
Query: 136 HPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKEASIVRFDVSGDSFFMAVDEIVNVHQ 195
G + D A + +R R S DK + ++ SF A D+I
Sbjct: 595 TRRGPPLVVSGSDDGTAKLWDMRQRGSIQTFPDK------YQITAVSFSDASDKI----- 741
Query: 196 AEDARLLMELQCPKRVLCAAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKG 255
YTGG D ++ WDL+ G+V + + H +
Sbjct: 742 ---------------------------YTGGIDNDVKIWDLRKGEVTMTL-QGHQDMITS 837
Query: 256 IVVLSDEATGDDEPYLVASASSDGTIRAWDVRMAATE 292
+ + D YL+ + D + WD+R A +
Sbjct: 838 MQL------SPDGSYLLTN-GMDCKLCIWDMRPYAPQ 927
Score = 30.8 bits (68), Expect = 0.74
Identities = 20/75 (26%), Positives = 33/75 (43%)
Frame = +1
Query: 222 LYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTI 281
+ + D+ + WD ++GK + E H + V P LV S S DGT
Sbjct: 487 IISASPDKTLRLWDTETGKQIKKMVE-HLSYVNSCCPTRRG------PPLVVSGSDDGTA 645
Query: 282 RAWDVRMAATEKSEP 296
+ WD+R + ++ P
Sbjct: 646 KLWDMRQRGSIQTFP 690
>TC86503 similar to GP|15028127|gb|AAK76687.1 unknown protein {Arabidopsis
thaliana}, partial (54%)
Length = 1915
Score = 42.4 bits (98), Expect = 2e-04
Identities = 24/95 (25%), Positives = 45/95 (47%)
Frame = +1
Query: 28 QTLTPVFSYPSHLSLIKTVAVSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALS 87
+T+T P + + N+++A G D +I +Y++ L H+ +T L+
Sbjct: 1261 KTMTTFMPPPPAATFLAFHPQDNNIIAIGMDDSSIQIYNVRVDEVKSKLKGHTKRITGLA 1440
Query: 88 FYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKT 122
F N+ LVS+ AD + +++ DG+ KT
Sbjct: 1441 FSHVLNV-----LVSSGADAQICVWNTDGWEKQKT 1530
Score = 32.0 bits (71), Expect = 0.33
Identities = 22/69 (31%), Positives = 29/69 (41%)
Frame = +3
Query: 216 PAKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASA 275
P K + T G+D+ I WD SG Y E H A V + E + S
Sbjct: 63 PNKQLCVITCGDDKTIKVWDAVSGAKQYTF-EGHEAPVYSVCPHYKE-----NIQFIFST 224
Query: 276 SSDGTIRAW 284
+ DG I+AW
Sbjct: 225 ALDGKIKAW 251
>TC77774 similar to GP|15028127|gb|AAK76687.1 unknown protein {Arabidopsis
thaliana}, partial (31%)
Length = 1487
Score = 42.0 bits (97), Expect = 3e-04
Identities = 23/90 (25%), Positives = 44/90 (48%)
Frame = +3
Query: 28 QTLTPVFSYPSHLSLIKTVAVSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALS 87
+T+T P + + N+++A G D +I +Y++ L H+ +T L+
Sbjct: 324 KTMTTFMPPPPAATFLAFHPQDNNIIAIGMDDSSIQIYNVRVDEVKSKLKGHTKRITGLA 503
Query: 88 FYSPPNLPFPRNLVSADADGSLAIFDADGF 117
F N+ LVS+ ADG + +++ DG+
Sbjct: 504 FSHVLNV-----LVSSGADGQIFVWNTDGW 578
>AL383150 similar to GP|17529236|gb putative SecA-type chloroplast protein
transport factor {Arabidopsis thaliana}, partial (14%)
Length = 445
Score = 42.0 bits (97), Expect = 3e-04
Identities = 25/70 (35%), Positives = 34/70 (47%)
Frame = +3
Query: 217 AKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASAS 276
A LLYTG DR I AW LK G + C + H + V + V + ++ S S
Sbjct: 171 AFGNLLYTGSGDRTIKAWSLKDGTL-MCTMDGHKSVVSTLSVCDE---------VLYSGS 320
Query: 277 SDGTIRAWDV 286
DGT+R W +
Sbjct: 321 WDGTVRLWSL 350
>TC82910 similar to GP|19347844|gb|AAL86002.1 unknown protein {Arabidopsis
thaliana}, partial (19%)
Length = 821
Score = 42.0 bits (97), Expect = 3e-04
Identities = 45/181 (24%), Positives = 75/181 (40%), Gaps = 3/181 (1%)
Frame = +1
Query: 117 FVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGR-RSFCCRLDKEASIVR 175
F+ + + +G+ LAI+ + IALT S D +VN+ GR S V
Sbjct: 13 FMLCEVIHTIPRGITCLAINSTSTIALTGSVDGSVHIVNITTGRVVSTLPSHSSSIECVG 192
Query: 176 FDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVLCAAPAKNGLLYTGGEDRNITAWD 235
F SG + + + + E + + V C+ + TG D + WD
Sbjct: 193 FAPSGSWAAIGGMDKMTIWDVEHSLARSICEHEYGVTCSTWLGTSYVATGSNDGAVRLWD 372
Query: 236 LKSGKVAYCIE--EAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAWDVRMAATEK 293
+SG+ C+ H+ ++ + + ++ YLV SAS D T R +DV+ K
Sbjct: 373 SRSGE---CVRTFRGHSESIQSL------SLSANQEYLV-SASLDHTARVFDVKGVC*AK 522
Query: 294 S 294
S
Sbjct: 523 S 525
Score = 29.6 bits (65), Expect = 1.6
Identities = 9/40 (22%), Positives = 25/40 (62%)
Frame = +1
Query: 49 SNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSF 88
++++ +G D ++H+ +++T + +L HSS++ + F
Sbjct: 76 TSTIALTGSVDGSVHIVNITTGRVVSTLPSHSSSIECVGF 195
>TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein beta
subunit-like protein. [Alfalfa] {Medicago sativa},
complete
Length = 1261
Score = 40.4 bits (93), Expect = 0.001
Identities = 44/201 (21%), Positives = 89/201 (43%), Gaps = 4/201 (1%)
Frame = +2
Query: 10 VAGSYERFIWGFNLNPNTQTLTPVFSYPSHLSLIKTVAVSNSVVASGGSDDTIHLYDL-- 67
++GS++ + ++LN T V LS+ ++ N + S D TI L++
Sbjct: 296 LSGSWDGELRLWDLNAGTSARRFVGHTKDVLSV--AFSIDNRQIVSASRDRTIKLWNTLG 469
Query: 68 STSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHK 127
++ HS V+ + F SP L +VSA D ++ +++ TL+ H
Sbjct: 470 ECKYTIQDGDAHSDWVSCVRF-SPSTLQ--PTIVSASWDRTVKVWNLTNCKLRNTLAGHS 640
Query: 128 KGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKEASIVRFDVSGDSFFM-- 185
VN +A+ P G + + +D + +L G+R + LD + I S + +++
Sbjct: 641 GYVNTVAVSPDGSLCASGGKDGVILLWDLAEGKRLY--SLDAGSIIHALCFSPNRYWLCA 814
Query: 186 AVDEIVNVHQAEDARLLMELQ 206
A + + + E ++ +L+
Sbjct: 815 ATESSIKIWDLESKSIVEDLK 877
Score = 32.0 bits (71), Expect = 0.33
Identities = 50/227 (22%), Positives = 85/227 (37%), Gaps = 18/227 (7%)
Frame = +2
Query: 38 SHLSLIKTVAV---SNSVVASGGSDDTIHLYDLSTSSSLGS-----LTDHSSTVTALSFY 89
+H ++ +A ++ ++ + D +I L+ L+ LT HS V +
Sbjct: 98 AHTDVVTAIATPIDNSDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFVQDVVLS 277
Query: 90 SPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDS 149
S +S DG L ++D + + H K V +A + ++ SRD
Sbjct: 278 SDGQFA-----LSGSWDGELRLWDLNAGTSARRFVGHTKDVLSVAFSIDNRQIVSASRDR 442
Query: 150 CFAMVNLVRGRRSFCCRLDKEA-----SIVRFDVSG---DSFFMAVDEIVNVHQAEDARL 201
+ N + G + + D +A S VRF S + D V V + +L
Sbjct: 443 TIKLWNTL-GECKYTIQ-DGDAHSDWVSCVRFSPSTLQPTIVSASWDRTVKVWNLTNCKL 616
Query: 202 LMELQCPKRVL--CAAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIE 246
L + A L +GG+D I WDL GK Y ++
Sbjct: 617 RNTLAGHSGYVNTVAVSPDGSLCASGGKDGVILLWDLAEGKRLYSLD 757
>TC82717 weakly similar to GP|21539583|gb|AAM53344.1 putative coatomer
complex subunit {Arabidopsis thaliana}, partial (29%)
Length = 1118
Score = 39.7 bits (91), Expect = 0.002
Identities = 24/99 (24%), Positives = 45/99 (45%)
Frame = +1
Query: 51 SVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLA 110
S AS D T+ ++ + +S+ + H + + ++ + + L+S D +
Sbjct: 556 STFASASLDGTLKIWTIDSSAPNFTFEGHLKGMNCVDYFESNDKQY---LLSGSDDYTAK 726
Query: 111 IFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDS 149
++D D ++TL HK V + HP I +T S DS
Sbjct: 727 VWDYDSKNCVQTLEGHKNNVTAICAHPEIPIIITASEDS 843
Score = 28.5 bits (62), Expect = 3.7
Identities = 18/68 (26%), Positives = 30/68 (43%)
Frame = +1
Query: 223 YTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIR 282
Y +D+ + W+ K G E ++ V + A +P ASAS DGT++
Sbjct: 433 YXASDDQVLKLWNWKKGWSCDETFEGNSHYVMQV------AFNPKDPSTFASASLDGTLK 594
Query: 283 AWDVRMAA 290
W + +A
Sbjct: 595 IWTIDSSA 618
Score = 28.1 bits (61), Expect = 4.8
Identities = 18/77 (23%), Positives = 33/77 (42%)
Frame = +3
Query: 61 TIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHL 120
TI +++ T + SL S V A F N +++A D + +++ + +
Sbjct: 201 TISIWNYQTKTEEKSLKVSESPVRAAKFIVRENW-----IIAATDDKYIRVYNYEKMEKI 365
Query: 121 KTLSVHKKGVNDLAIHP 137
HK + LA+HP
Sbjct: 366 IEFEAHKDYIRSLAVHP 416
>TC79362 WD-repeat cell cycle regulatory protein
Length = 1800
Score = 39.3 bits (90), Expect = 0.002
Identities = 42/153 (27%), Positives = 66/153 (42%), Gaps = 7/153 (4%)
Frame = +3
Query: 17 FIWGFNLNPNTQTLTPVFSYPSHLSLIKTVAVS---NSVVASGG--SDDTIHLYDLSTSS 71
F+W N + PV Y H + +K +A S + ++ASGG +D I ++ +T+S
Sbjct: 1113 FVW------NQHSTQPVLKYCEHTAAVKAIAWSPHLHGLLASGGGTADRCIRFWNTTTNS 1274
Query: 72 SLGSLTDHSSTVTALSFYSPPNLPFPRNLVSAD--ADGSLAIFDADGFVHLKTLSVHKKG 129
L S D S V L + N LVS + + ++ L TL+ H
Sbjct: 1275 HL-SCMDTGSQVCNLVWSKNVN-----ELVSTHGYSQNQIIVWRYPTMSKLATLTGHTYR 1436
Query: 130 VNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRS 162
V LAI P G+ +T + D N+ +S
Sbjct: 1437 VLYLAISPDGQTIVTGAGDETLTFWNVFPSPKS 1535
>TC78145 similar to GP|15294240|gb|AAK95297.1 AT3g18860/MCB22_3 {Arabidopsis
thaliana}, partial (93%)
Length = 2633
Score = 38.9 bits (89), Expect = 0.003
Identities = 50/240 (20%), Positives = 86/240 (35%), Gaps = 3/240 (1%)
Frame = +3
Query: 50 NSVVASGGSDDTIHLYDLSTSSSLGS--LTDHSSTVTALSFYSPPNLPFPRN-LVSADAD 106
N +A+ D TI L+ + L H+S V +++ PPN P +VS D
Sbjct: 180 NGGIATSSRDKTIRLWTRDNRKFVLDKVLVGHTSFVGPIAWI-PPNPDLPHGGVVSGGMD 356
Query: 107 GSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCR 166
+ ++D + +++L H+ V + + ++ ++ L R R C
Sbjct: 357 TLVLVWDLNSGEKVQSLKGHQLQVTGIVVDDGDLVSSSMD-------CTLRRWRNGQC-- 509
Query: 167 LDKEASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVLCAAPAKNGLLYTGG 226
+ +A + ++ P G L TG
Sbjct: 510 ------------------------IETWEAHKGPIQAVIKLP----------TGELVTGS 587
Query: 227 EDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAWDV 286
D + W GK + HA V+G+ V+SD + SAS DG++R W V
Sbjct: 588 SDTTLKTW---KGKTCLHTFQGHADTVRGLAVMSDLG--------ILSASHDGSLRLWAV 734
>TC90372 similar to GP|20197166|gb|AAC27402.2 expressed protein {Arabidopsis
thaliana}, partial (52%)
Length = 706
Score = 38.9 bits (89), Expect = 0.003
Identities = 21/64 (32%), Positives = 32/64 (49%)
Frame = +1
Query: 224 TGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRA 283
TG D +I A D+++G ++ AH A V ++ L++ VAS DG I+
Sbjct: 298 TGSPDFSILATDVETGSTIARLDNAHEAAVNRLINLTEST--------VASGDDDGCIKV 453
Query: 284 WDVR 287
WD R
Sbjct: 454 WDTR 465
>CB893690 weakly similar to GP|9759025|dbj gene_id:K6A12.9~similar to unknown
protein~sp|O15736 {Arabidopsis thaliana}, partial (59%)
Length = 916
Score = 37.4 bits (85), Expect = 0.008
Identities = 30/122 (24%), Positives = 51/122 (41%)
Frame = +3
Query: 28 QTLTPVFSYPSHLSLIKTVAVSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALS 87
QT + + + + VS+ +V S D TI ++DL ++ S AL
Sbjct: 465 QTTYTLTGHTDKVCAVDISKVSSRLVVSAAYDGTIKVWDLMKGYCTNTIM-FPSICNALC 641
Query: 88 FYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSR 147
F + F S DG+L ++D L ++ H V +++ +G IALT
Sbjct: 642 FSTDGQTIF-----SGHVDGNLRLWDIQSGRLLSEVATHSHAVTSISLSRNGNIALTSGG 806
Query: 148 DS 149
D+
Sbjct: 807 DN 812
Score = 31.2 bits (69), Expect(2) = 0.004
Identities = 22/88 (25%), Positives = 37/88 (42%)
Frame = +2
Query: 191 VNVHQAEDARLLMELQCPKRVLCAAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHA 250
+N H+ A LL E K L TGG+D ++ WD +G ++ +
Sbjct: 263 LNAHEGGCASLLFEYNSSK------------LITGGQDHSVKVWDTNTGSLSSSLTGCLG 406
Query: 251 ARVKGIVVLSDEATGDDEPYLVASASSD 278
+ VL T D+ + AS+S++
Sbjct: 407 S------VLDLTITNDNRSVIAASSSNN 472
Score = 26.2 bits (56), Expect(2) = 0.004
Identities = 10/16 (62%), Positives = 13/16 (80%)
Frame = +3
Query: 271 LVASASSDGTIRAWDV 286
LV SA+ DGTI+ WD+
Sbjct: 537 LVVSAAYDGTIKVWDL 584
>BG647724 similar to GP|11141605|gb LEUNIG {Arabidopsis thaliana}, partial
(25%)
Length = 757
Score = 37.0 bits (84), Expect = 0.010
Identities = 24/88 (27%), Positives = 44/88 (49%), Gaps = 1/88 (1%)
Frame = +2
Query: 52 VVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAI 111
++ASGG D + L+ + +L +HS +T + F P++P L ++ D ++ +
Sbjct: 473 LLASGGHDKKVVLWYTDSLKQKATLEEHSLLITDVRF--SPSMP---RLATSSYDKTVRV 637
Query: 112 FDADG-FVHLKTLSVHKKGVNDLAIHPS 138
+D D L+T + H V L HP+
Sbjct: 638 WDVDNPGYSLRTFTGHSAAVMSLDFHPN 721
Score = 35.0 bits (79), Expect = 0.039
Identities = 25/78 (32%), Positives = 42/78 (53%), Gaps = 1/78 (1%)
Frame = +2
Query: 210 RVLCAAPAKNG-LLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDE 268
+V+C+ + +G LL +GG D+ + W S K +EE H+ ++++D
Sbjct: 437 KVVCSHFSSDGKLLASGGHDKKVVLWYTDSLKQKATLEE-HS------LLITDVRFSPSM 595
Query: 269 PYLVASASSDGTIRAWDV 286
P L A++S D T+R WDV
Sbjct: 596 PRL-ATSSYDKTVRVWDV 646
>TC86168 homologue to GP|13936812|gb|AAK49947.1 TGF-beta
receptor-interacting protein 1 {Phaseolus vulgaris},
complete
Length = 1322
Score = 36.6 bits (83), Expect = 0.013
Identities = 49/210 (23%), Positives = 80/210 (37%), Gaps = 15/210 (7%)
Frame = +2
Query: 100 LVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRG 159
L S D + ++ AD L T H V + +T S D + N+ G
Sbjct: 242 LFSCAKDHNPTVWFADNGERLGTYRGHNGAVWCCDVSRDSGRLITGSADQTAKLWNVQTG 421
Query: 160 RRSFCCRLDKEASIVRFDVSGDSF-------FMAVDEIVNVHQA------EDARLLMELQ 206
++ F D A V F V GD FM + ++V + + A L+ ++
Sbjct: 422 QQLFTFNFDSPARSVDFSV-GDKLAVITTDPFMELSSAIHVKRIAKDPSDQTAESLLVIK 598
Query: 207 CPKRVLCAA--PAKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEAT 264
P+ + A N + + GED I WD ++GK+ + K I L+ A
Sbjct: 599 GPQGRINRAIWGPLNKTIISAGEDAVIRIWDSETGKLLK-ESDKELGHKKTITSLAKSAD 775
Query: 265 GDDEPYLVASASSDGTIRAWDVRMAATEKS 294
G + S D + + WD R + K+
Sbjct: 776 GSH----FCTGSLDKSAKIWDSRTLSLVKT 853
>BF004489 similar to GP|20161344|d contains ESTs AU086067(S4642)
D41819(S4642)~similar to Arabidopsis thaliana chromosome
1 At1g49040~, partial (16%)
Length = 605
Score = 36.6 bits (83), Expect = 0.013
Identities = 24/80 (30%), Positives = 41/80 (51%), Gaps = 2/80 (2%)
Frame = +1
Query: 210 RVLCAAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIEE--AHAARVKGIVVLSDEATGDD 267
R + A + G + +G +D+++ WD ++ ++ +EE H V + LS E
Sbjct: 28 RTVRAISSDMGKVVSGSDDQSVLVWDKQTTQL---LEELKGHDGPVSCVRTLSGER---- 186
Query: 268 EPYLVASASSDGTIRAWDVR 287
V +AS DGT++ WDVR
Sbjct: 187 ----VLTASHDGTVKMWDVR 234
>TC92298 similar to PIR|T00642|T00642 hypothetical protein F3I6.5 -
Arabidopsis thaliana, partial (38%)
Length = 881
Score = 35.8 bits (81), Expect = 0.023
Identities = 23/90 (25%), Positives = 39/90 (42%), Gaps = 10/90 (11%)
Frame = +3
Query: 209 KRVLCAAPAKNGLLYTGGEDRNITAWDLKSGKVAY---CIEEAHAARVKGIVVLSDEATG 265
K +LC + L+ +G D ++ W + +Y + + H VK + + D +G
Sbjct: 501 KAILCLV-VMDDLVCSGSADNSVRLWKRGIDEKSYTCLAVLQGHRKAVKCLAIADDSKSG 677
Query: 266 -------DDEPYLVASASSDGTIRAWDVRM 288
D YLV S S D I+ W +R+
Sbjct: 678 KNGGVDDDGSSYLVYSGSLDCDIKVWQIRV 767
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,136,493
Number of Sequences: 36976
Number of extensions: 136789
Number of successful extensions: 861
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 800
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 850
length of query: 324
length of database: 9,014,727
effective HSP length: 96
effective length of query: 228
effective length of database: 5,465,031
effective search space: 1246027068
effective search space used: 1246027068
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144656.10


