

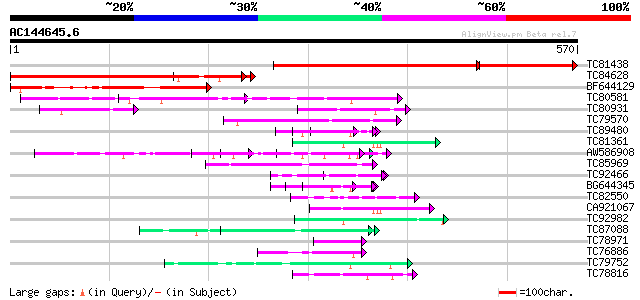
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144645.6 - phase: 0 /pseudo
(570 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81438 similar to GP|20259253|gb|AAM14362.1 unknown protein {Ar... 408 e-167
TC84628 similar to GP|20259253|gb|AAM14362.1 unknown protein {Ar... 376 e-104
BF644129 168 5e-42
TC80581 similar to GP|19683021|gb|AAL92644.1 hypothetical protei... 140 1e-33
TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem tra... 69 6e-12
TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (AR... 59 4e-09
TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Ar... 59 7e-09
TC81361 similar to GP|6143870|gb|AAF04417.1| unknown protein {Ar... 58 1e-08
AW586908 weakly similar to GP|2104814|emb| pleiotropic effects o... 55 7e-08
TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsi... 54 2e-07
TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis... 51 1e-06
BG644345 weakly similar to GP|19697333|gb putative protein poten... 50 3e-06
TC82550 similar to GP|3832528|gb|AAC70787.1| unknown {Glycine ma... 49 5e-06
CA921067 similar to GP|6143870|gb|A unknown protein {Arabidopsis... 48 1e-05
TC92982 similar to GP|23332794|gb|AAN27343.1 Sequence 13 from pa... 46 3e-05
TC87088 similar to PIR|F86399|F86399 protein F17L21.22 [imported... 45 8e-05
TC78971 similar to GP|20260346|gb|AAM13071.1 unknown protein {Ar... 44 1e-04
TC76886 similar to PIR|T01932|T01932 RNA binding protein homolog... 44 2e-04
TC79752 similar to GP|11994670|dbj|BAB02898. gene_id:K7P8.17~unk... 44 2e-04
TC78816 similar to GP|14190377|gb|AAK55669.1 AT4g00850/A_TM018A1... 43 3e-04
>TC81438 similar to GP|20259253|gb|AAM14362.1 unknown protein {Arabidopsis
thaliana}, partial (21%)
Length = 1334
Score = 408 bits (1049), Expect(2) = e-167
Identities = 206/209 (98%), Positives = 207/209 (98%)
Frame = +2
Query: 266 MHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQ 325
MHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQ
Sbjct: 2 MHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQ 181
Query: 326 QKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLT 385
QKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLT
Sbjct: 182 QKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLT 361
Query: 386 GSQPDATASGATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFA 445
GSQPDATASGATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFA
Sbjct: 362 GSQPDATASGATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFA 541
Query: 446 DDFIDSVTTHGCILAKHRKSSTLESKDLL 474
DDFIDSVTTHGCILAKH KSSTLESKD +
Sbjct: 542 DDFIDSVTTHGCILAKHXKSSTLESKDFI 628
Score = 198 bits (503), Expect(2) = e-167
Identities = 98/98 (100%), Positives = 98/98 (100%)
Frame = +3
Query: 473 LLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVRMLMESSSVPESIVNNSKDI 532
LLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVRMLMESSSVPESIVNNSKDI
Sbjct: 624 LLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVRMLMESSSVPESIVNNSKDI 803
Query: 533 SRQGHPNPAGSHHLMRPLSSDQLVSHSTSSQMLQQMTR 570
SRQGHPNPAGSHHLMRPLSSDQLVSHSTSSQMLQQMTR
Sbjct: 804 SRQGHPNPAGSHHLMRPLSSDQLVSHSTSSQMLQQMTR 917
Score = 33.1 bits (74), Expect = 0.30
Identities = 44/147 (29%), Positives = 60/147 (39%), Gaps = 6/147 (4%)
Frame = +2
Query: 14 QNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQ 73
Q P+ S P+ SP+ QQ SS L Q+ L+ QL Q +Q
Sbjct: 38 QMPAMSGPA----SPLRLQQHQRQQQQLASSGQLQQNSMTLN---------QQQLSQFMQ 178
Query: 74 RSPSMSR--LNQIQP----QQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAAM 127
+ SM + L+Q QP QQQQQ++ QQ + A+V QQ S VA
Sbjct: 179 QQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQL--------QASVHQQQHLHSPRVAGP 334
Query: 128 GGSASNLSRSALIGQSGHFPMLSGAGA 154
G S I +G P + +GA
Sbjct: 335 TGQKS-------ISLTGSQPDATASGA 394
>TC84628 similar to GP|20259253|gb|AAM14362.1 unknown protein {Arabidopsis
thaliana}, partial (8%)
Length = 839
Score = 376 bits (966), Expect = e-104
Identities = 205/243 (84%), Positives = 210/243 (86%), Gaps = 5/243 (2%)
Frame = +1
Query: 1 MAENVIGSPDSQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTI 60
MAENVIGSPDSQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTI
Sbjct: 94 MAENVIGSPDSQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTI 273
Query: 61 NPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQL 120
NPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQL
Sbjct: 274 NPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQL 453
Query: 121 SGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGG-----MVQSSQF 175
SGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGG + +F
Sbjct: 454 SGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGDGAILTIFFGEF 633
Query: 176 SSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNS 235
+A + G P + TNGGLYTQQQQIRLTPAQMRQQLSQQALNS
Sbjct: 634 CWTITARNATSYWDDG----GRPI*PLK*GTNGGLYTQQQQIRLTPAQMRQQLSQQALNS 801
Query: 236 QQV 238
QQV
Sbjct: 802 QQV 810
Score = 64.7 bits (156), Expect = 9e-11
Identities = 47/88 (53%), Positives = 55/88 (62%), Gaps = 5/88 (5%)
Frame = +2
Query: 165 QKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGG---LYTQQQQIRLTP 221
+K GMVQSSQFSSANSAGQSLQGMQQAIGMMG +A +N G ++T R
Sbjct: 587 KKVGMVQSSQFSSANSAGQSLQGMQQAIGMMG---VAQSSLSNEGPMEVFTLSSS-RFD* 754
Query: 222 AQMRQQLSQQA--LNSQQVQGIPRSSSL 247
Q + S + L + QGIPRSSSL
Sbjct: 755 LQPK*GSSSHSRHLTVNRFQGIPRSSSL 838
Score = 33.1 bits (74), Expect = 0.30
Identities = 44/147 (29%), Positives = 60/147 (39%), Gaps = 28/147 (19%)
Frame = +1
Query: 278 QMPAMSGPA----SPLRLQQHQRQQQQLASSGQLQQNSMTLN---------QQQLSQFMQ 324
Q P+ S P+ SP+ QQ SS L Q+ L+ QL Q +Q
Sbjct: 133 QNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQ 312
Query: 325 QQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQL--------QASVHQQQHLHSPRVAGP 376
+ SM + L+Q QP QQQQQ++ QQ + A+V QQ S VA
Sbjct: 313 RSPSMSR--LNQIQP----QQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAAM 474
Query: 377 TGQKS-------ISLTGSQPDATASGA 396
G S I +G P + +GA
Sbjct: 475 GGSASNLSRSALIGQSGHFPMLSGAGA 555
>BF644129
Length = 511
Score = 168 bits (426), Expect = 5e-42
Identities = 117/208 (56%), Positives = 127/208 (60%), Gaps = 5/208 (2%)
Frame = +1
Query: 1 MAENVIGSP----DSQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHT 56
MA+N+I S D+Q QNP SSNP+IPSPSPI QQ P+I N SP
Sbjct: 34 MAQNMISSQQSTIDTQTQNPISSNPTIPSPSPIPQQL--PAIINLNPSP----------- 174
Query: 57 INTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQ 116
NPN PS QIQP QQQ VA QQA LYGG MNFGGSAAV Q
Sbjct: 175 ----NPNV-----------PSF----QIQPLQQQ--VAVQQAGLYGGLMNFGGSAAVPDQ 291
Query: 117 QQQLSGGVAA-MGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQF 175
QQQLSG V +GG+ASNLS GAGA QFNLLTSP QK G+VQ+SQ
Sbjct: 292 QQQLSGSVGVGVGGNASNLS---------------GAGATQFNLLTSPGQKVGLVQTSQL 426
Query: 176 SSANSAGQSLQGMQQAIGMMGSPNLASQ 203
SSANSAGQSLQGMQQAIG +GS NLASQ
Sbjct: 427 SSANSAGQSLQGMQQAIGTLGSSNLASQ 510
>TC80581 similar to GP|19683021|gb|AAL92644.1 hypothetical protein
{Dictyostelium discoideum}, partial (5%)
Length = 850
Score = 140 bits (353), Expect = 1e-33
Identities = 123/303 (40%), Positives = 154/303 (50%), Gaps = 17/303 (5%)
Frame = +1
Query: 110 SAAVSAQQQQLSGGVAAMGGSASNLSRSALIG--QSGHFPMLSG---------AGAAQFN 158
+ A+ AQQ QLS + + L+G QSG M+ +G+A
Sbjct: 22 NGALYAQQLQLS--------QQAPFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSASLL 177
Query: 159 LLTSPRQKGGMVQSSQF--SSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQ 216
L P+ + + S+Q SS Q L + Q MG P L Q++ L QQQ
Sbjct: 178 PLQQPQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLL---QQQ 348
Query: 217 IRLTPAQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWL 276
++L Q Q+L QQ +S Q+Q +SL QL L Q QP M L Q Q
Sbjct: 349 LQLQ--QQYQRLQQQLASSAQLQ----QNSLTLNQQQLPLLMQ--QPKSMGQPLLQQQQQ 504
Query: 277 KQMPAMSGPASPLRLQQ-HQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLH 335
+ + L+LQQ +QR QQQLASS QLQQNS TLNQ L Q MQQQKSMGQP L
Sbjct: 505 QLLQQQ------LQLQQQYQRLQQQLASSAQLQQNSSTLNQ--LPQLMQQQKSMGQPLLQ 660
Query: 336 QQQPSPQ---QQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDAT 392
QQ Q QQQQ L Q QQQ Q Q + QQ + + R+ GP G KS+SLTGSQP+AT
Sbjct: 661 QQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQALRIPGPAGHKSLSLTGSQPEAT 840
Query: 393 ASG 395
A G
Sbjct: 841 AFG 849
Score = 57.0 bits (136), Expect = 2e-08
Identities = 68/234 (29%), Positives = 98/234 (41%), Gaps = 4/234 (1%)
Frame = +1
Query: 12 QIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQT 71
QI S S +P P SQQQ + S + SS +L+Q QQ + P QLQQ
Sbjct: 142 QIPAVSGSASLLPLQQPQSQQQLASSAQLQQSSLTLNQ-QQLPQLMQQPKPMGQPQLQQQ 318
Query: 72 LQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQ---LSGGVAAMG 128
LQ + + Q+Q QQQ Q + +Q A+ Q N +++ QQQ L +MG
Sbjct: 319 LQHQQLLQQ--QLQLQQQYQRLQQQLASSAQLQQN-----SLTLNQQQLPLLMQQPKSMG 477
Query: 129 GSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTS-PRQKGGMVQSSQFSSANSAGQSLQG 187
+ L+ Q Q L R + + S+Q +S L
Sbjct: 478 QPLLQQQQQQLLQQ-------------QLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQ 618
Query: 188 MQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQGI 241
+ Q MG P L Q++ L QQQQ+ L Q +QQ + L QQ+Q +
Sbjct: 619 LMQQQKSMGQPLLQQQLQQQ-QLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQAL 777
>TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem transcription
factor M1 {Apium graveolens}, partial (67%)
Length = 1193
Score = 68.6 bits (166), Expect = 6e-12
Identities = 50/118 (42%), Positives = 63/118 (53%), Gaps = 4/118 (3%)
Frame = +2
Query: 290 RLQQHQRQQQQLASSGQLQQNS-MTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQ 348
+LQQ +QQQQ QLQQN ++ QQQ+ Q QQQ+ + PQL QQQ S QQQQQQ
Sbjct: 563 QLQQQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQL--PQLQQQQLSQLQQQQQQ 736
Query: 349 LLQPQQQSQLQASVHQQQ---HLHSPRVAGPTGQKSISLTGSQPDATASGATTPGGSS 403
L Q QQ Q QQ + V GP G+ + SQ ++ GAT GG +
Sbjct: 737 LPQLQQLQHQQLPQQQQMVGAGMGQTYVQGPGGRSQMV---SQGQVSSQGATNIGGGN 901
Score = 42.4 bits (98), Expect = 5e-04
Identities = 31/103 (30%), Positives = 44/103 (42%), Gaps = 4/103 (3%)
Frame = +2
Query: 31 QQQQSPSIHMSNSSPSLSQD----QQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQP 86
QQQQ P + + + Q QQQ + + QLQQ Q+ P + +L Q
Sbjct: 593 QQQQLPQLQQNQQLSQIQQQIPQLQQQQQQLPQLQQQQLSQLQQQQQQLPQLQQLQHQQL 772
Query: 87 QQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAAMGG 129
QQQQ+V Y GG + + +Q Q S G +GG
Sbjct: 773 PQQQQMVGAGMGQTY--VQGPGGRSQMVSQGQVSSQGATNIGG 895
Score = 39.3 bits (90), Expect = 0.004
Identities = 33/122 (27%), Positives = 54/122 (44%), Gaps = 6/122 (4%)
Frame = +1
Query: 295 QRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQ 354
Q Q+++L + QL ++ L+ + + G + + Q S QQQQQQ +Q QQ
Sbjct: 343 QLQEKKLCAVIQLPSQTLLLSVSDKACRLIGMLFPGDMVVFKPQLSGQQQQQQ--MQQQQ 516
Query: 355 QSQLQASVHQQQHL------HSPRVAGPTGQKSISLTGSQPDATASGATTPGGSSSQGTE 408
Q Q QQQHL + T + + + + PD+TA T +++ T
Sbjct: 517 MQQHQQMQSQQQHLSTIAATDATTTTTATASTAATESAAFPDSTADSTTPATAAATPSTS 696
Query: 409 AA 410
A
Sbjct: 697 TA 702
Score = 34.3 bits (77), Expect = 0.13
Identities = 38/120 (31%), Positives = 45/120 (36%), Gaps = 4/120 (3%)
Frame = +2
Query: 198 PNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALN----SQQVQGIPRSSSLAFMNSQ 253
P L QM+ QQQQ +L Q QQLSQ QQ Q +P+ Q
Sbjct: 560 PQLQQQMQQ------QQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQLPQ-----LQQQQ 706
Query: 254 LSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMT 313
LS L Q Q L Q Q L Q M G Q + Q+ S GQ+ T
Sbjct: 707 LSQLQQQQQQLPQLQQL-QHQQLPQQQQMVGAGMGQTYVQGPGGRSQMVSQGQVSSQGAT 883
Score = 34.3 bits (77), Expect = 0.13
Identities = 67/273 (24%), Positives = 96/273 (34%), Gaps = 4/273 (1%)
Frame = +1
Query: 142 QSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLA 201
QSG P+ + A AA L SSQ SL G +Q G P
Sbjct: 25 QSGMQPL*NNAAAANMTLTQQ-------TASSQSKYIKVWEGSLSGQRQ-----GQPVFI 168
Query: 202 SQMRTNGGLYTQQQQIRLTPAQMR--QQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQ 259
+++ + P M+ + +SQ +N++Q G MN
Sbjct: 169 TKLEGYRSSSASETLAANWPPVMQIVRLISQDHMNNKQYVGKADFLVFRAMNP------- 327
Query: 260 NGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQL 319
H L Q Q K + P+ L L + + + G L M + + QL
Sbjct: 328 -------HGFLGQLQEKKLCAVIQLPSQTLLLSVSDKACRLI---GMLFPGDMVVFKPQL 477
Query: 320 SQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQ 379
S QQQ+ M Q QQ QQ Q +Q QQQ L + A
Sbjct: 478 SG-QQQQQQMQQ----------QQMQQHQQMQSQQQ-HLSTIAATDATTTTTATASTAAT 621
Query: 380 KSISLTGSQPDAT--ASGATTPGGSSSQGTEAA 410
+S + S D+T A+ A TP S++ AA
Sbjct: 622 ESAAFPDSTADSTTPATAAATPSTSTAAAFSAA 720
Score = 30.4 bits (67), Expect = 1.9
Identities = 39/142 (27%), Positives = 55/142 (38%), Gaps = 7/142 (4%)
Frame = +2
Query: 43 SSPSLSQDQQQLHTINTINPNSNF--QLQQTLQRSPSMSRLNQIQP-----QQQQQVVAR 95
SS S + + + + N NS+ QLQQ +Q+ +L Q+Q Q QQQ+
Sbjct: 485 SSSSNRCNSNKCNNTSRCNRNSSIFPQLQQQMQQQQQQQQLPQLQQNQQLSQIQQQIPQL 664
Query: 96 QQAALYGGQMNFGGSAAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAA 155
QQ Q+ + + QQQQL L L Q + GAG
Sbjct: 665 QQQQQQLPQLQQQQLSQLQQQQQQLP--------QLQQLQHQQLPQQQ----QMVGAGMG 808
Query: 156 QFNLLTSPRQKGGMVQSSQFSS 177
Q + P + MV Q SS
Sbjct: 809 Q-TYVQGPGGRSQMVSQGQVSS 871
Score = 30.0 bits (66), Expect = 2.5
Identities = 29/100 (29%), Positives = 43/100 (43%), Gaps = 6/100 (6%)
Frame = +2
Query: 49 QDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVAR-QQAALYGGQMNF 107
Q QQQL + N QL Q Q+ P + + Q PQ QQQ +++ QQ Q+
Sbjct: 590 QQQQQLPQLQ-----QNQQLSQIQQQIPQLQQQQQQLPQLQQQQLSQLQQQQQQLPQLQQ 754
Query: 108 GGSAAVSAQQQQLSGG-----VAAMGGSASNLSRSALIGQ 142
+ QQQ + G V GG + +S+ + Q
Sbjct: 755 LQHQQLPQQQQMVGAGMGQTYVQGPGGRSQMVSQGQVSSQ 874
>TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (15%)
Length = 1143
Score = 59.3 bits (142), Expect = 4e-09
Identities = 54/186 (29%), Positives = 77/186 (41%), Gaps = 7/186 (3%)
Frame = +2
Query: 216 QIRLTPAQMRQQ------LSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNS 269
Q RL P+ M Q ++ AL + + + + Q NG +M N
Sbjct: 254 QARLDPSMMNMQPDMYQAMAAAALQDMRTLDPSKQHPASLLQFQQPQNFSNGNAALMQNQ 433
Query: 270 -LTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKS 328
L QSQ + P P + Q +Q QQL N QQQ Q QQQ+
Sbjct: 434 MLQQSQPHQAFPKNQESQHPSQSQAQTQQFQQLLQHQHSFTNQNYHMQQQQQQ--QQQQQ 607
Query: 329 MGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQ 388
Q Q HQQQ Q QQQQQ++ QQ S +++ Q + +P ++IS G Q
Sbjct: 608 QQQQQQHQQQQQQQSQQQQQVVDHQQMSNAVSTM--SQFVSAPPSQSTQPMQAISSIGHQ 781
Query: 389 PDATAS 394
+ + S
Sbjct: 782 QNFSDS 799
Score = 40.4 bits (93), Expect = 0.002
Identities = 42/147 (28%), Positives = 60/147 (40%), Gaps = 7/147 (4%)
Frame = +2
Query: 12 QIQNPSSSNPSIPSPSPISQQQ-------QSPSIHMSNSSPSLSQDQQQLHTINTINPNS 64
Q QN S+ N ++ + Q Q S H S S Q QQ L ++ N
Sbjct: 386 QPQNFSNGNAALMQNQMLQQSQPHQAFPKNQESQHPSQSQAQTQQFQQLLQHQHSFT-NQ 562
Query: 65 NFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGV 124
N+ +QQ Q+ + Q Q QQQQQ ++QQ + Q S AVS Q +S
Sbjct: 563 NYHMQQQQQQQQQQQQQQQQQHQQQQQQQSQQQQQVVDHQQM---SNAVSTMSQFVS--- 724
Query: 125 AAMGGSASNLSRSALIGQSGHFPMLSG 151
A S + + IG +F +G
Sbjct: 725 APPSQSTQPMQAISSIGHQQNFSDSNG 805
Score = 32.7 bits (73), Expect = 0.39
Identities = 79/334 (23%), Positives = 115/334 (33%), Gaps = 17/334 (5%)
Frame = +2
Query: 16 PSSSNPSIPSPSPISQQQQ----SPSIH--------MSNSSPSLSQDQQQLHTINTINPN 63
P ++ P PSP P+ ++ PS H M++ L + L T+N
Sbjct: 59 PLTTFPMYPSPFPLRLKRPWPPGLPSFHGLKDDDFGMNSPLLWLRDADRGLQTLNFQGMG 238
Query: 64 SNFQLQQTLQRSPSMSRLNQIQPQQQQQVVAR--QQAALYGGQMNFGGSAAVSAQQQQLS 121
N +Q L PSM + QP Q + A Q S Q Q S
Sbjct: 239 VNPWMQARLD--PSMMNM---QPDMYQAMAAAALQDMRTLDPSKQHPASLLQFQQPQNFS 403
Query: 122 GGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSA 181
G AA L ++ ++ QS P Q Q SQ S + A
Sbjct: 404 NGNAA-------LMQNQMLQQS------------------QPHQAFPKNQESQHPSQSQA 508
Query: 182 G-QSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQG 240
Q Q + Q + N Q + QQQQ + Q +QQ QQ+ QQV
Sbjct: 509 QTQQFQQLLQHQHSFTNQNYHMQQQQQ----QQQQQQQQQQQQHQQQQQQQSQQQQQVVD 676
Query: 241 IPRSSSLAFMNSQLSGL--SQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQ 298
+ S+ SQ SQ+ QP +S+ Q + P SPL
Sbjct: 677 HQQMSNAVSTMSQFVSAPPSQSTQPMQAISSIGHQQNFSD--SNGNPVSPL--------- 823
Query: 299 QQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQP 332
+ S + S LN + + ++ Q S +P
Sbjct: 824 HNMLGSFSNDETSHLLNFPRPNSWVPVQSSTARP 925
>TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 1068
Score = 58.5 bits (140), Expect = 7e-09
Identities = 38/85 (44%), Positives = 47/85 (54%)
Frame = +1
Query: 285 PASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQ 344
P S ++Q + Q + LQQ L+QQQ F Q Q + Q Q QQQ QQQ
Sbjct: 97 PNSNPQIQSQSQSQSNQDPNLYLQQQQQFLHQQQQQPFQQSQTTQSQFQQQQQQQLYQQQ 276
Query: 345 QQQQLLQPQQQSQLQASVHQQQHLH 369
QQQ++LQ QQQ QLQ QQQ+LH
Sbjct: 277 QQQRILQ-QQQQQLQ-QPQQQQNLH 345
Score = 46.2 bits (108), Expect = 3e-05
Identities = 32/81 (39%), Positives = 42/81 (51%), Gaps = 11/81 (13%)
Frame = +1
Query: 303 SSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSP-----------QQQQQQQLLQ 351
S+ Q+Q S + + Q + ++QQQ Q LHQQQ P QQQQQQQL Q
Sbjct: 103 SNPQIQSQSQSQSNQDPNLYLQQQ----QQFLHQQQQQPFQQSQTTQSQFQQQQQQQLYQ 270
Query: 352 PQQQSQLQASVHQQQHLHSPR 372
QQQ ++ QQQ L P+
Sbjct: 271 QQQQQRILQ--QQQQQLQQPQ 327
Score = 42.7 bits (99), Expect = 4e-04
Identities = 35/86 (40%), Positives = 39/86 (44%), Gaps = 3/86 (3%)
Frame = +1
Query: 268 NSLTQSQWLKQMPAMSGPASPLRLQQ---HQRQQQQLASSGQLQQNSMTLNQQQLSQFMQ 324
NS Q Q Q + P L+ QQ HQ+QQQ S Q QQQL Q Q
Sbjct: 100 NSNPQIQSQSQSQSNQDPNLYLQQQQQFLHQQQQQPFQQSQTTQSQFQQQQQQQLYQQQQ 279
Query: 325 QQKSMGQPQLHQQQPSPQQQQQQQLL 350
QQ+ L QQQ QQ QQQQ L
Sbjct: 280 QQRI-----LQQQQQQLQQPQQQQNL 342
Score = 40.0 bits (92), Expect = 0.002
Identities = 29/84 (34%), Positives = 39/84 (45%)
Frame = +1
Query: 18 SSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPS 77
+SNP I S S SQ Q P++++ L Q QQQ + S FQ QQ Q
Sbjct: 100 NSNPQIQSQSQ-SQSNQDPNLYLQQQQQFLHQQQQQPFQ-QSQTTQSQFQQQQQQQLYQQ 273
Query: 78 MSRLNQIQPQQQQQVVARQQAALY 101
+ +Q QQQQ +QQ L+
Sbjct: 274 QQQQRILQQQQQQLQQPQQQQNLH 345
Score = 39.7 bits (91), Expect = 0.003
Identities = 30/85 (35%), Positives = 40/85 (46%), Gaps = 2/85 (2%)
Frame = +1
Query: 15 NPSSSNPSIPSPSPISQQQQSPSIHMSNSSPS--LSQDQQQLHTINTINPNSNFQLQQTL 72
+P + IP+P+ + Q QS S SN P+ L Q QQ LH FQ QT
Sbjct: 64 SPGGTWTMIPTPNS-NPQIQSQSQSQSNQDPNLYLQQQQQFLHQ----QQQQPFQQSQTT 228
Query: 73 QRSPSMSRLNQIQPQQQQQVVARQQ 97
Q + Q+ QQQQQ + +QQ
Sbjct: 229 QSQFQQQQQQQLYQQQQQQRILQQQ 303
Score = 29.3 bits (64), Expect = 4.3
Identities = 26/85 (30%), Positives = 36/85 (41%), Gaps = 3/85 (3%)
Frame = +1
Query: 11 SQIQNPSSSNPSI---PSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQ 67
SQ Q+ S+ +P++ + QQQQ P S ++ S Q QQQ
Sbjct: 121 SQSQSQSNQDPNLYLQQQQQFLHQQQQQP-FQQSQTTQSQFQQQQQ----------QQLY 267
Query: 68 LQQTLQRSPSMSRLNQIQPQQQQQV 92
QQ QR + QPQQQQ +
Sbjct: 268 QQQQQQRILQQQQQQLQQPQQQQNL 342
Score = 28.1 bits (61), Expect = 9.5
Identities = 22/85 (25%), Positives = 34/85 (39%)
Frame = +1
Query: 186 QGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQGIPRSS 245
Q QQ + Q + LY QQQQ R+ Q +QQL Q Q +
Sbjct: 190 QQQQQPFQQSQTTQSQFQQQQQQQLYQQQQQQRILQ-QQQQQLQQPQQQQNLHQSLASHF 366
Query: 246 SLAFMNSQLSGLSQNGQPGMMHNSL 270
L + L+ + ++G P ++L
Sbjct: 367 HLLNLVENLAEVVEHGNPDQQSDAL 441
>TC81361 similar to GP|6143870|gb|AAF04417.1| unknown protein {Arabidopsis
thaliana}, partial (9%)
Length = 668
Score = 57.8 bits (138), Expect = 1e-08
Identities = 53/179 (29%), Positives = 72/179 (39%), Gaps = 30/179 (16%)
Frame = +3
Query: 285 PASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQL----HQQQPS 340
PA Q H + A +S + Q ++Q G+P L ++QQ +
Sbjct: 63 PAGSQSFQGHGLMRPSSAGPPSAPSSSASQGMQSINQPWLSSGPPGKPPLPSPAYRQQIN 242
Query: 341 PQQQQQQQLLQPQQQSQLQASVHQ--------QQHL----------HSP------RVAGP 376
PQ QQ+ + QQQS AS Q Q+H H P R+ GP
Sbjct: 243 PQSLQQRTHISQQQQSMPTASQQQQPLPSNQTQEHFGQQVPSSRAPHVPHQAQVTRLQGP 422
Query: 377 TGQKSISLTGSQPDATASGATT--PGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKL 433
QK SL Q A G+ + P + E + VL KR I +LV QVDP GK+
Sbjct: 423 GNQKPSSLVAGQSGAVQPGSQSRLPNTLPNADIEESGKSVLSKRSIHELVHQVDPLGKV 599
Score = 29.3 bits (64), Expect = 4.3
Identities = 24/94 (25%), Positives = 38/94 (39%), Gaps = 11/94 (11%)
Frame = +3
Query: 24 PSPSPISQQQQSPS-----IHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSP-- 76
P PSP +QQ +P H+S S+ QQ + + +F Q R+P
Sbjct: 207 PLPSPAYRQQINPQSLQQRTHISQQQQSMPTASQQQQPLPSNQTQEHFGQQVPSSRAPHV 386
Query: 77 ----SMSRLNQIQPQQQQQVVARQQAALYGGQMN 106
++RL Q+ +VA Q A+ G +
Sbjct: 387 PHQAQVTRLQGPGNQKPSSLVAGQSGAVQPGSQS 488
>AW586908 weakly similar to GP|2104814|emb| pleiotropic effects on cellular
differentiation and slug behaviour {Dictyostelium
discoideum}, partial (8%)
Length = 668
Score = 55.1 bits (131), Expect = 7e-08
Identities = 68/194 (35%), Positives = 80/194 (41%), Gaps = 20/194 (10%)
Frame = +2
Query: 183 QSLQGMQQAIGMMGSPNLASQ---MRTNGGLYTQQQQIRLTPAQ-------------MRQ 226
Q Q QQ G P Q + QQQQ P Q +
Sbjct: 89 QQQQRQQQLAGSQNLPQPQQQQPQLTQQSPQQPQQQQQHQQPCQNTTMNNGTIGSNQIPN 268
Query: 227 QLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQ-SQWLKQMPAMSGP 285
Q QQ + Q+Q S S M SQ L G+ G+M SL Q SQ+ +Q+
Sbjct: 269 QCVQQPVTYSQLQQQLLSGS---MQSQ-QNLQSAGKNGLMMTSLPQDSQFQQQIDQQQ-- 430
Query: 286 ASPLRLQQHQRQQQQLASSGQLQQNSMTLN---QQQLSQFMQQQKSMGQPQLHQQQPSPQ 342
A L+ QQ Q Q QQ SS QL Q SM Q Q+SQ + Q S Q QL Q Q
Sbjct: 431 AGLLQRQQQQNQLQQ--SSLQLLQQSMLQRGPQQPQMSQTVPQNISDQQQQLQLLQKLQQ 604
Query: 343 QQQQQQLLQPQQQS 356
QQQQQQ QP S
Sbjct: 605 QQQQQQQQQPLSTS 646
Score = 50.1 bits (118), Expect = 2e-06
Identities = 68/234 (29%), Positives = 95/234 (40%), Gaps = 14/234 (5%)
Frame = +2
Query: 26 PSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQ 85
P+ SQQQQ S L Q QQQ + + N L Q Q+ P +++ + Q
Sbjct: 11 PASWSQQQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQN--LPQPQQQQPQLTQQSPQQ 184
Query: 86 PQQQQQVVARQQAALYGGQMNFG--GSAAV------------SAQQQQLSGGVAAMGGSA 131
PQQQQQ Q MN G GS + QQQ LSG + S
Sbjct: 185 PQQQQQ----HQQPCQNTTMNNGTIGSNQIPNQCVQQPVTYSQLQQQLLSGSMQ----SQ 340
Query: 132 SNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSAGQSLQGMQQA 191
NL + G++G M S +QF +Q G + + Q + + SLQ +QQ+
Sbjct: 341 QNLQSA---GKNG-LMMTSLPQDSQFQQQIDQQQAGLLQRQQQQNQLQQS--SLQLLQQS 502
Query: 192 IGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQGIPRSS 245
+ G + QQQQ+ Q+ Q+L QQ QQ Q + SS
Sbjct: 503 MLQRGPQQPQMSQTVPQNISDQQQQL-----QLLQKLQQQQQQQQQQQPLSTSS 649
Score = 49.7 bits (117), Expect = 3e-06
Identities = 52/165 (31%), Positives = 65/165 (38%), Gaps = 11/165 (6%)
Frame = +2
Query: 213 QQQQIRLTPAQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLS-QNGQPGMMHNSLT 271
QQQ+++ QL QQ QQ+ G QL+ S Q Q H
Sbjct: 38 QQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQLTQQSPQQPQQQQQHQQPC 217
Query: 272 QSQWLKQMPAMSGPASPLRLQQ----HQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQK 327
Q+ + S +QQ Q QQQ L+ S Q QQN + + L Q
Sbjct: 218 QNTTMNNGTIGSNQIPNQCVQQPVTYSQLQQQLLSGSMQSQQNLQSAGKNGLMMTSLPQD 397
Query: 328 SMGQPQLHQQQPSPQQQQQQQ------LLQPQQQSQLQASVHQQQ 366
S Q Q+ QQQ Q+QQQQ LQ QQS LQ Q Q
Sbjct: 398 SQFQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRGPQQPQ 532
Score = 45.4 bits (106), Expect = 6e-05
Identities = 46/132 (34%), Positives = 54/132 (40%), Gaps = 16/132 (12%)
Frame = +2
Query: 269 SLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKS 328
S +Q Q +Q P +LQQ Q++QQQLA S L Q QQQ Q QQ
Sbjct: 17 SWSQQQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQP-----QQQQPQLTQQSPQ 181
Query: 329 MGQPQLHQQQPS---------------PQQQQQQQLLQPQ-QQSQLQASVHQQQHLHSPR 372
Q Q QQP P Q QQ + Q QQ L S+ QQ+L S
Sbjct: 182 QPQQQQQHQQPCQNTTMNNGTIGSNQIPNQCVQQPVTYSQLQQQLLSGSMQSQQNLQS-- 355
Query: 373 VAGPTGQKSISL 384
AG G SL
Sbjct: 356 -AGKNGLMMTSL 388
Score = 41.2 bits (95), Expect = 0.001
Identities = 33/95 (34%), Positives = 39/95 (40%)
Frame = +2
Query: 288 PLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQ 347
P Q Q+QQQ++ S L L Q QQQ+ Q Q P PQQQQ Q
Sbjct: 11 PASWSQQQQQQQKMQS----------LLPMPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQ 160
Query: 348 QLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSI 382
Q QQ Q Q HQQ ++ G G I
Sbjct: 161 LTQQSPQQPQ-QQQQHQQPCQNTTMNNGTIGSNQI 262
Score = 41.2 bits (95), Expect = 0.001
Identities = 32/76 (42%), Positives = 39/76 (51%), Gaps = 2/76 (2%)
Frame = +2
Query: 285 PASPLRLQQHQRQQQQLASS--GQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQ 342
PAS + QQ Q++ Q L QLQQ Q SQ + Q + QPQL QQ P Q
Sbjct: 11 PASWSQQQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQPQQQ-QPQLTQQSPQ-Q 184
Query: 343 QQQQQQLLQPQQQSQL 358
QQQQQ QP Q + +
Sbjct: 185 PQQQQQHQQPCQNTTM 232
Score = 29.3 bits (64), Expect = 4.3
Identities = 22/59 (37%), Positives = 27/59 (45%), Gaps = 5/59 (8%)
Frame = +2
Query: 336 QQQPSP--QQQQQQQLLQ---PQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQP 389
QQ P+ QQQQQQQ +Q P QLQ +QQ L + Q+ LT P
Sbjct: 2 QQPPASWSQQQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQLTQQSP 178
Score = 28.5 bits (62), Expect = 7.3
Identities = 19/51 (37%), Positives = 23/51 (44%), Gaps = 5/51 (9%)
Frame = +2
Query: 331 QPQLHQQQPSPQQQQQQQLL-----QPQQQSQLQASVHQQQHLHSPRVAGP 376
QP Q QQQ+ Q LL Q QQQ Q Q + Q+L P+ P
Sbjct: 5 QPPASWSQQQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQPQQQQP 157
Score = 28.1 bits (61), Expect = 9.5
Identities = 26/68 (38%), Positives = 30/68 (43%), Gaps = 16/68 (23%)
Frame = +2
Query: 316 QQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQ----------LLQPQQQS-QL-----Q 359
QQ + + QQQ+ + Q P Q QQQQQ L QPQQQ QL Q
Sbjct: 2 QQPPASWSQQQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQLTQQSPQ 181
Query: 360 ASVHQQQH 367
QQQH
Sbjct: 182 QPQQQQQH 205
>TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsis thaliana},
partial (31%)
Length = 2756
Score = 53.9 bits (128), Expect = 2e-07
Identities = 48/172 (27%), Positives = 71/172 (40%)
Frame = +2
Query: 198 PNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGL 257
P +Q +T +Q Q P Q +Q+ S + + SSL+ S+ +
Sbjct: 1310 PQAQNQAQTQSQAQSQAQTQSQAP-QFQQKSSGGGSDLPSPDSVSSDSSLSNAMSRQKPV 1486
Query: 258 SQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQ 317
Q + + S + +S P S +++QQH + G L Q L QQ
Sbjct: 1487 VYQEQIQIQPGTTRVSNPVDPKLNLSDPHSRIQVQQH------VQDPGYLLQQQFELQQQ 1648
Query: 318 QLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLH 369
Q +QQQ+ Q Q Q QP Q QQQQQL Q Q Q +H ++H
Sbjct: 1649 QQQFELQQQQQQPQHQPQQSQPQHQPQQQQQL----QHHQQQQFIHGGHYIH 1792
Score = 41.2 bits (95), Expect = 0.001
Identities = 49/211 (23%), Positives = 74/211 (34%), Gaps = 9/211 (4%)
Frame = +2
Query: 165 QKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQM 224
Q QS S A + Q+ Q Q++ G G +L S + R P
Sbjct: 1319 QNQAQTQSQAQSQAQTQSQAPQFQQKSSG--GGSDLPSPDSVSSDSSLSNAMSRQKPVVY 1492
Query: 225 RQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSG 284
++Q+ Q ++ + +L+ +S++ PG + L+Q +
Sbjct: 1493 QEQIQIQPGTTRVSNPVDPKLNLSDPHSRIQVQQHVQDPGYL---------LQQQFELQQ 1645
Query: 285 PASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQ------- 337
LQQ Q+Q Q Q Q QQQL QQQ G +H
Sbjct: 1646 QQQQFELQQQQQQPQHQPQQSQPQHQPQ--QQQQLQHHQQQQFIHGGHYIHHNPAIPTYY 1819
Query: 338 --QPSPQQQQQQQLLQPQQQSQLQASVHQQQ 366
PS QQ Q P +Q Q ++ QQ
Sbjct: 1820 PVYPSQQQPHHQVYYVPARQPQQGYNISVQQ 1912
Score = 40.0 bits (92), Expect = 0.002
Identities = 45/163 (27%), Positives = 60/163 (36%), Gaps = 3/163 (1%)
Frame = +2
Query: 220 TPAQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQM 279
TP Q+ QA N Q Q ++ S A SQ Q G +
Sbjct: 1280 TPQPQVVQVQPQAQNQAQTQS--QAQSQAQTQSQAPQFQQKSSGGG-----------SDL 1420
Query: 280 PAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTL---NQQQLSQFMQQQKSMGQPQLHQ 336
P+ +S L +Q+ + Q+Q T N + S Q Q H
Sbjct: 1421 PSPDSVSSDSSLSNAMSRQKPVVYQEQIQIQPGTTRVSNPVDPKLNLSDPHSRIQVQQHV 1600
Query: 337 QQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQ 379
Q P QQQ +L Q QQQ +LQ QQQ H P+ + P Q
Sbjct: 1601 QDPGYLLQQQFELQQQQQQFELQQQ--QQQPQHQPQQSQPQHQ 1723
Score = 37.0 bits (84), Expect = 0.021
Identities = 58/226 (25%), Positives = 87/226 (37%), Gaps = 17/226 (7%)
Frame = +2
Query: 222 AQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPA 281
AQ +QQL QQ Q+V GI A M + G+ Q Q +
Sbjct: 992 AQQQQQLQQQQ-QDQKVLGI--DEQFAQMVVGVGGIGQRLQD-------------EGFAV 1123
Query: 282 MSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKS-MGQPQLHQQQPS 340
MS P H LA+ G +++ + Q F ++S G P +++ P+
Sbjct: 1124 MSSPP-------HPPVPTTLAAVGVPIGSAVVVGDYQNRVFSDDERSDHGVPVGYRKPPT 1282
Query: 341 PQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGS---QPDATASGAT 397
PQ Q Q +QPQ Q+Q Q Q + A P Q+ S GS PD+ +S ++
Sbjct: 1283 PQPQVVQ--VQPQAQNQAQTQSQAQSQAQTQSQA-PQFQQKSSGGGSDLPSPDSVSSDSS 1453
Query: 398 TPGGSSSQ-------------GTEAATNQVLGKRKIQDLVAQVDPQ 430
S Q GT +N V K + D +++ Q
Sbjct: 1454 LSNAMSRQKPVVYQEQIQIQPGTTRVSNPVDPKLNLSDPHSRIQVQ 1591
Score = 31.6 bits (70), Expect = 0.86
Identities = 35/132 (26%), Positives = 50/132 (37%), Gaps = 42/132 (31%)
Frame = +2
Query: 14 QNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHT------------INTIN 61
Q S +PSP +S S S MS P + Q+Q Q+ +N +
Sbjct: 1391 QKSSGGGSDLPSPDSVSSDS-SLSNAMSRQKPVVYQEQIQIQPGTTRVSNPVDPKLNLSD 1567
Query: 62 PNSNFQLQQTLQ-----------------------------RSPSMSRLNQIQPQQQQQV 92
P+S Q+QQ +Q P S+ Q QPQQQQQ+
Sbjct: 1568 PHSRIQVQQHVQDPGYLLQQQFELQQQQQQFELQQQQQQPQHQPQQSQ-PQHQPQQQQQL 1744
Query: 93 V-ARQQAALYGG 103
+QQ ++GG
Sbjct: 1745 QHHQQQQFIHGG 1780
>TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis thaliana},
partial (14%)
Length = 632
Score = 51.2 bits (121), Expect = 1e-06
Identities = 40/116 (34%), Positives = 56/116 (47%)
Frame = +1
Query: 263 PGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQF 322
PG+M ++L +WL + ++ L+ ++ Q+QQQQ Q QQ+ QQQ Q
Sbjct: 301 PGLMKSTL---RWLHHI*RLN*----LKAREQQQQQQQQQQQPQPQQSQHAQQQQQ--QH 453
Query: 323 MQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTG 378
MQ Q+ + Q QQQ Q QQQ Q QPQQ Q + HL + G G
Sbjct: 454 MQMQQLLMQRHAQQQQQQQQHQQQPQ-SQPQQPQPQQQQNRDRTHLLNGSANGLAG 618
Score = 45.1 bits (105), Expect = 8e-05
Identities = 27/66 (40%), Positives = 31/66 (46%)
Frame = +1
Query: 316 QQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAG 375
QQQ Q QQQ Q Q QQQ Q QQ L+Q Q Q Q HQQQ P+
Sbjct: 373 QQQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQQPQ 552
Query: 376 PTGQKS 381
P Q++
Sbjct: 553 PQQQQN 570
Score = 33.1 bits (74), Expect = 0.30
Identities = 25/76 (32%), Positives = 31/76 (39%)
Frame = +1
Query: 31 QQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQ 90
QQQQ P S + Q Q+ + Q QQ Q+ P S+ Q QPQQQQ
Sbjct: 391 QQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQ-SQPQQPQPQQQQ 567
Query: 91 QVVARQQAALYGGQMN 106
R + L G N
Sbjct: 568 N---RDRTHLLNGSAN 606
>BG644345 weakly similar to GP|19697333|gb putative protein potential
transcriptional repressor Not4hp - Mus musculus, partial
(7%)
Length = 635
Score = 49.7 bits (117), Expect = 3e-06
Identities = 39/107 (36%), Positives = 45/107 (41%)
Frame = +1
Query: 263 PGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQF 322
PG S Q+ +Q P QQQ + Q+ S QQQ QF
Sbjct: 169 PGQQQASPFQTPGQQQSSPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQ---QQQQLQF 339
Query: 323 MQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLH 369
QQQ+ Q QQQ QQQQQ QL Q QQQ Q QQQ L+
Sbjct: 340 QQQQQQQQQQLQQQQQQQLQQQQQLQLQQQQQQLLQQHQSSQQQQLY 480
Score = 44.7 bits (104), Expect = 1e-04
Identities = 40/87 (45%), Positives = 41/87 (46%), Gaps = 13/87 (14%)
Frame = +1
Query: 295 QRQQQQLASSGQLQQNS--MTLNQQQLSQFM---QQQKSMGQPQLHQQQPSP-------- 341
Q QQ L + QQ S T QQQ S F QQQ S Q QQQPSP
Sbjct: 139 QAQQPSLFQTPGQQQASPFQTPGQQQSSPFQTPGQQQASPFQTP-GQQQPSPFSQITPFS 315
Query: 342 QQQQQQQLLQPQQQSQLQASVHQQQHL 368
QQQQQ Q Q QQQ Q Q QQQ L
Sbjct: 316 QQQQQLQFQQQQQQQQQQLQQQQQQQL 396
Score = 44.3 bits (103), Expect = 1e-04
Identities = 38/87 (43%), Positives = 41/87 (46%), Gaps = 11/87 (12%)
Frame = +1
Query: 295 QRQQQQLASSGQLQQNSM-TLNQQQLSQF--------MQQQKSMGQPQLHQQQPSPQQQQ 345
Q+Q + GQ Q + T QQQ S F QQQ Q Q QQQ QQQQ
Sbjct: 208 QQQSSPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQQQQQLQFQQQQQQQQQQLQQQQQ 387
Query: 346 QQQLLQPQQQSQLQASVHQ--QQHLHS 370
QQ LQ QQQ QLQ Q QQH S
Sbjct: 388 QQ--LQQQQQLQLQQQQQQLLQQHQSS 462
Score = 43.5 bits (101), Expect = 2e-04
Identities = 29/74 (39%), Positives = 34/74 (45%), Gaps = 2/74 (2%)
Frame = +1
Query: 278 QMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQ- 336
Q P P+ ++ +QQQQL Q QQ L QQQ Q QQQ+ Q Q Q
Sbjct: 262 QTPGQQQPSPFSQITPFSQQQQQLQFQQQQQQQQQQLQQQQQQQLQQQQQLQLQQQQQQL 441
Query: 337 -QQPSPQQQQQQQL 349
QQ QQQQ L
Sbjct: 442 LQQHQSSQQQQLYL 483
Score = 40.8 bits (94), Expect = 0.001
Identities = 40/115 (34%), Positives = 50/115 (42%), Gaps = 6/115 (5%)
Frame = +1
Query: 12 QIQNPSS-SNPSIPSPSPIS---QQQQSP--SIHMSNSSPSLSQDQQQLHTINTINPNSN 65
Q Q PS P SP QQQ SP + +SP + QQQ + I P S
Sbjct: 139 QAQQPSLFQTPGQQQASPFQTPGQQQSSPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQ 318
Query: 66 FQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQL 120
Q Q Q+ + Q+Q QQQQQ+ +QQ L Q S+QQQQL
Sbjct: 319 QQQQLQFQQQQQQQQ-QQLQQQQQQQLQQQQQLQLQQQQQQL-LQQHQSSQQQQL 477
Score = 33.9 bits (76), Expect = 0.17
Identities = 36/115 (31%), Positives = 45/115 (38%), Gaps = 5/115 (4%)
Frame = +1
Query: 226 QQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGP 285
Q QQ + Q G +SS Q + Q PG Q Q+ S
Sbjct: 163 QTPGQQQASPFQTPGQQQSSPFQTPGQQQASPFQT--PGQQ-----QPSPFSQITPFSQQ 321
Query: 286 ASPLRLQQHQRQ-----QQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLH 335
L+ QQ Q+Q QQQ QLQQ QQQ Q +QQ +S Q QL+
Sbjct: 322 QQQLQFQQQQQQQQQQLQQQ--QQQQLQQQQQLQLQQQQQQLLQQHQSSQQQQLY 480
Score = 32.0 bits (71), Expect = 0.66
Identities = 29/79 (36%), Positives = 35/79 (43%), Gaps = 1/79 (1%)
Frame = +1
Query: 162 SPRQKGGMVQSSQFSSANSAGQSLQGMQ-QAIGMMGSPNLASQMRTNGGLYTQQQQIRLT 220
SP Q G Q S FS Q Q +Q Q L Q + QQQQ++L
Sbjct: 253 SPFQTPGQQQPSPFSQITPFSQQQQQLQFQQQQQQQQQQLQQQQQQQ---LQQQQQLQLQ 423
Query: 221 PAQMRQQLSQQALNSQQVQ 239
Q +QQL QQ +SQQ Q
Sbjct: 424 --QQQQQLLQQHQSSQQQQ 474
Score = 30.8 bits (68), Expect = 1.5
Identities = 32/107 (29%), Positives = 40/107 (36%), Gaps = 2/107 (1%)
Frame = +1
Query: 16 PSSSNPSIPSPSPISQQQQSP--SIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQ 73
P + PS+ QQQ SP + SSP + QQQ T Q
Sbjct: 136 PQAQQPSLFQTP--GQQQASPFQTPGQQQSSPFQTPGQQQASPFQTPGQQQPSPFSQITP 309
Query: 74 RSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQL 120
S +L Q QQQQQ +QQ Q+ + QQQQL
Sbjct: 310 FSQQQQQLQFQQQQQQQQQQLQQQQQ---QQLQQQQQLQLQQQQQQL 441
>TC82550 similar to GP|3832528|gb|AAC70787.1| unknown {Glycine max}, partial
(32%)
Length = 875
Score = 48.9 bits (115), Expect = 5e-06
Identities = 45/130 (34%), Positives = 59/130 (44%)
Frame = +1
Query: 283 SGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQ 342
S P P Q+ Q+QQQQ LQQ S++ N QQ S G PQ +Q S Q
Sbjct: 34 SNPHLPSLQQRQQQQQQQ-----HLQQRSLSANN-----LPQQNHSQG-PQGNQ---SLQ 171
Query: 343 QQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATASGATTPGGS 402
QQ QQLLQ S V QQ H P V+G + ++ G P T +G + G
Sbjct: 172 QQMIQQLLQ--DMSNNNGGVQQQSH-SGPNVSGNMAKNNLGFGGQTP--TTNGGGSANGQ 336
Query: 403 SSQGTEAATN 412
+ G + +N
Sbjct: 337 GNNGPVSRSN 366
Score = 35.0 bits (79), Expect = 0.078
Identities = 42/136 (30%), Positives = 53/136 (38%), Gaps = 9/136 (6%)
Frame = +1
Query: 19 SNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTL--QRSP 76
SNP +PS QQQQ H+ S S + QQ H + P N LQQ + Q
Sbjct: 34 SNPHLPSLQQRQQQQQQQ--HLQQRSLSANNLPQQNH---SQGPQGNQSLQQQMIQQLLQ 198
Query: 77 SMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAV-----SAQQQQLSGGVAAMGG-- 129
MS N QQQ + + FGG SA Q +G V+
Sbjct: 199 DMSN-NNGGVQQQSHSGPNVSGNMAKNNLGFGGQTPTTNGGGSANGQGNNGPVSRSNSFK 375
Query: 130 SASNLSRSALIGQSGH 145
+ASN SA G G+
Sbjct: 376 AASNSDSSAAAGGGGN 423
>CA921067 similar to GP|6143870|gb|A unknown protein {Arabidopsis thaliana},
partial (10%)
Length = 727
Score = 47.8 bits (112), Expect = 1e-05
Identities = 46/152 (30%), Positives = 62/152 (40%), Gaps = 26/152 (17%)
Frame = -1
Query: 302 ASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQAS 361
A S Q ++NQ LS + + P ++QQ +PQ QQ+ + QQQS AS
Sbjct: 664 APSSSASQGMQSINQPWLSSGPPGKPPLPSPA-YRQQINPQSLQQRTHISQQQQSMPTAS 488
Query: 362 VHQ--------QQHL----------HSP------RVAGPTGQKSISLTGSQPDATASGAT 397
Q Q+H H P R+ GP QK SL Q A G+
Sbjct: 487 QQQQPLPSNQTQEHFGQQVPSSRAPHVPHQAQVTRLQGPGNQKPSSLVAGQSGAVQPGSQ 308
Query: 398 T--PGGSSSQGTEAATNQVLGKRKIQDLVAQV 427
+ P + E + VL KR I +LV QV
Sbjct: 307 SRLPNTLPNADIEESGKSVLSKRSIHELVHQV 212
Score = 29.3 bits (64), Expect = 4.3
Identities = 24/94 (25%), Positives = 38/94 (39%), Gaps = 11/94 (11%)
Frame = -1
Query: 24 PSPSPISQQQQSPS-----IHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSP-- 76
P PSP +QQ +P H+S S+ QQ + + +F Q R+P
Sbjct: 586 PLPSPAYRQQINPQSLQQRTHISQQQQSMPTASQQQQPLPSNQTQEHFGQQVPSSRAPHV 407
Query: 77 ----SMSRLNQIQPQQQQQVVARQQAALYGGQMN 106
++RL Q+ +VA Q A+ G +
Sbjct: 406 PHQAQVTRLQGPGNQKPSSLVAGQSGAVQPGSQS 305
>TC92982 similar to GP|23332794|gb|AAN27343.1 Sequence 13 from patent US
6440698, partial (1%)
Length = 795
Score = 46.2 bits (108), Expect = 3e-05
Identities = 44/170 (25%), Positives = 62/170 (35%), Gaps = 15/170 (8%)
Frame = -1
Query: 287 SPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQL------HQQQPS 340
SP R + Q A+S Q +S T Q Q + S Q + QQ +
Sbjct: 780 SPKRNTSYNNAASQPAASSYSQPSSTTQQQTASLQSFNMRVSASASQSRSSSTKYSQQQA 601
Query: 341 PQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATASGATTPG 400
Q QQQQQ QPQQ Q Q + + + + P + P + SG + G
Sbjct: 600 QQAQQQQQQQQPQQTQQTQQTQSYGSYPPQAQPSHPEQHSWYGFGSNAPSSFTSGGNSAG 421
Query: 401 GSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKL---------DPEVIDLL 441
G S++ + A N G Q G L D ++ DLL
Sbjct: 420 GYSARSSTTA-NYGTGANSANQAYQQQQHSGSLNMSSHNYTGDNDIYDLL 274
Score = 32.7 bits (73), Expect = 0.39
Identities = 25/79 (31%), Positives = 36/79 (44%)
Frame = -1
Query: 161 TSPRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLT 220
TS +S +S +S Q Q+ M S + ASQ R++ Y+ QQQ +
Sbjct: 765 TSYNNAASQPAASSYSQPSSTTQQQTASLQSFNMRVSAS-ASQSRSSSTKYS-QQQAQQA 592
Query: 221 PAQMRQQLSQQALNSQQVQ 239
Q +QQ QQ +QQ Q
Sbjct: 591 QQQQQQQQPQQTQQTQQTQ 535
Score = 30.0 bits (66), Expect = 2.5
Identities = 44/166 (26%), Positives = 62/166 (36%)
Frame = -1
Query: 41 SNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAAL 100
S S PS S QQQ ++ + N + Q+ S S+ Q QQQQQ QQ
Sbjct: 726 SYSQPS-STTQQQTASLQSFNMRVSASASQSRSSSTKYSQQQAQQAQQQQQQQQPQQTQQ 550
Query: 101 YGGQMNFGGSAAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLL 160
++G + Q Q + G SN S G G A ++
Sbjct: 549 TQQTQSYG---SYPPQAQPSHPEQHSWYGFGSNAPSSFTSG---------GNSAGGYSAR 406
Query: 161 TSPRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRT 206
+S G + ANSA Q+ Q Q + GS N++S T
Sbjct: 405 SSTTANYG-------TGANSANQAYQQQQHS----GSLNMSSHNYT 301
>TC87088 similar to PIR|F86399|F86399 protein F17L21.22 [imported] -
Arabidopsis thaliana, partial (4%)
Length = 2997
Score = 45.1 bits (105), Expect = 8e-05
Identities = 57/242 (23%), Positives = 92/242 (37%), Gaps = 7/242 (2%)
Frame = +3
Query: 131 ASNLSRSALIGQSGHF--PMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSAGQSLQ-- 186
A + + S +G HF P++ T GGMV S F + +
Sbjct: 87 AQSSNSSLRLGDQSHFTDPLIDRDAP-----FTDQSSMGGMVNQSSFRDTWTDEYGINRH 251
Query: 187 -GMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQA--LNSQQVQGIPR 243
Q +G + L S++ N + + L Q + QQA L QQ + + +
Sbjct: 252 FNPNQRVGSLEDQFL-SRIGPNFNNFDVADHLMLQKLQKERLQQQQAERLQQQQAERLQQ 428
Query: 244 SSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLAS 303
+ Q L Q + + S + PA + R Q +
Sbjct: 429 QQAERLQQQQAERLQQQQAERLQQQTNISSHF----PAYLNGSDLDRFPGFSPSQSNKSG 596
Query: 304 SGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVH 363
Q+ QN T + ++L + QQ+ + ++ QQ QQ QQ LQPQQQSQ+Q +
Sbjct: 597 IQQMMQNPGT-DFERLFELQAQQRQL---EIQQQDMHHQQLLQQLKLQPQQQSQVQQLLL 764
Query: 364 QQ 365
+Q
Sbjct: 765 EQ 770
Score = 42.4 bits (98), Expect = 5e-04
Identities = 40/160 (25%), Positives = 60/160 (37%), Gaps = 1/160 (0%)
Frame = +3
Query: 213 QQQQIRLTPAQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQ 272
Q+++++ A+ QQ + L QQ + + + + Q L Q +
Sbjct: 360 QKERLQQQQAERLQQQQAERLQQQQAERLQQQQAERLQQQQAERLQQQTNISSHFPAYLN 539
Query: 273 SQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQP 332
L + P S S QQ + + G + L QQ +QQQ Q
Sbjct: 540 GSDLDRFPGFSPSQS-----NKSGIQQMMQNPGTDFERLFELQAQQRQLEIQQQDMHHQQ 704
Query: 333 QLHQQQPSPQQQQQ-QQLLQPQQQSQLQASVHQQQHLHSP 371
L Q + PQQQ Q QQLL Q Q + + Q H P
Sbjct: 705 LLQQLKLQPQQQSQVQQLLLEQLMHQQMSDPNFGQSKHDP 824
>TC78971 similar to GP|20260346|gb|AAM13071.1 unknown protein {Arabidopsis
thaliana}, partial (48%)
Length = 2260
Score = 44.3 bits (103), Expect = 1e-04
Identities = 27/53 (50%), Positives = 28/53 (51%)
Frame = +1
Query: 306 QLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQL 358
Q QQ QQQ QQQ S+ QPQ Q QP Q QQQQ Q QQQ QL
Sbjct: 262 QPQQTLFQPQQQQQQSPFQQQSSLFQPQQPQFQPQQQFQQQQFQAQQQQQQQL 420
Score = 38.1 bits (87), Expect = 0.009
Identities = 28/59 (47%), Positives = 29/59 (48%), Gaps = 1/59 (1%)
Frame = +1
Query: 292 QQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMG-QPQLHQQQPSPQQQQQQQL 349
QQ Q QQ Q QQ+ QQQ S F QQ Q Q QQQ QQQQQQQL
Sbjct: 253 QQSQPQQTLFQPQQQQQQSPF---QQQSSLFQPQQPQFQPQQQFQQQQFQAQQQQQQQL 420
Score = 36.2 bits (82), Expect = 0.035
Identities = 27/58 (46%), Positives = 30/58 (51%), Gaps = 10/58 (17%)
Frame = +1
Query: 321 QFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQ----------QQSQLQASVHQQQHL 368
Q Q Q+++ QPQ QQQ SP QQQ L QPQ QQ Q QA QQQ L
Sbjct: 253 QQSQPQQTLFQPQ-QQQQQSPFQQQSS-LFQPQQPQFQPQQQFQQQQFQAQQQQQQQL 420
Score = 28.9 bits (63), Expect = 5.6
Identities = 28/76 (36%), Positives = 29/76 (37%)
Frame = +1
Query: 16 PSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRS 75
P S P P QQQQSP S SL Q QQ FQ QQ Q+
Sbjct: 250 PQQSQPQQTLFQPQQQQQQSPFQQQS----SLFQPQQP-----------QFQPQQQFQQQ 384
Query: 76 PSMSRLNQIQPQQQQQ 91
Q Q QQQQQ
Sbjct: 385 -------QFQAQQQQQ 411
>TC76886 similar to PIR|T01932|T01932 RNA binding protein homolog - common
tobacco (fragment), partial (67%)
Length = 1896
Score = 43.9 bits (102), Expect = 2e-04
Identities = 40/125 (32%), Positives = 50/125 (40%), Gaps = 16/125 (12%)
Frame = +2
Query: 250 MNSQLSG--LSQN-GQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQ 306
M +Q +G L+QN Q H Q QW +Q P HQ QQQ + S Q
Sbjct: 203 MAAQTNGGDLNQNQNQNQNQHQQQQQQQWAQQQP-------------HQYQQQWM--SMQ 337
Query: 307 LQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQ-------------QQQQLLQPQ 353
+M + QQQ+ + Q P +QQQP QQQ QQ Q
Sbjct: 338 YPATAMAMMQQQMMMYPQHYMPYVHPHHYQQQPPHQQQVSPPPSAHSHHGGHQQHQYQQH 517
Query: 354 QQSQL 358
QQ QL
Sbjct: 518 QQKQL 532
Score = 37.0 bits (84), Expect = 0.021
Identities = 29/88 (32%), Positives = 38/88 (42%), Gaps = 9/88 (10%)
Frame = +2
Query: 303 SSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQ--------QQQQQLLQPQQ 354
+ G L QN NQ Q Q QQQ + QP +QQQ Q QQQ ++ PQ
Sbjct: 218 NGGDLNQNQ-NQNQNQHQQQQQQQWAQQQPHQYQQQWMSMQYPATAMAMMQQQMMMYPQH 394
Query: 355 -QSQLQASVHQQQHLHSPRVAGPTGQKS 381
+ +QQQ H +V+ P S
Sbjct: 395 YMPYVHPHHYQQQPPHQQQVSPPPSAHS 478
>TC79752 similar to GP|11994670|dbj|BAB02898. gene_id:K7P8.17~unknown
protein {Arabidopsis thaliana}, partial (2%)
Length = 2137
Score = 43.5 bits (101), Expect = 2e-04
Identities = 73/275 (26%), Positives = 95/275 (34%), Gaps = 25/275 (9%)
Frame = +1
Query: 156 QFNLLTSPRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQ 215
Q+N L S R M QSS + G + +GMMG N + M G
Sbjct: 211 QYNQLISSRN---MQQSSMSVPGSDRGARMLPGANGMGMMGGINRSIAMARPGFH----- 366
Query: 216 QIRLTPAQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQW 275
+T + M LS + S + G+P + NS + HN Q Q
Sbjct: 367 --GMTSSSM---LSSGGMLSSSMVGMPGVGA-GQGNSMMRPRDTVHMMRPGHNQGHQRQ- 525
Query: 276 LKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTL--NQQQLSQFMQQQKSMGQPQ 333
M P P+++ Q Q S NS T + QQ QQQ + P
Sbjct: 526 ------MMVPELPMQVTQGNSQGIPAFSGMSSAFNSQTTPPSVQQYPVHAQQQSHLSNPH 687
Query: 334 LHQQQPSP-----------------QQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGP 376
H Q P+ QQQQQQ+ LQ QQ S A + Q P ++ P
Sbjct: 688 PHLQGPNHPNNQQQAYIRLAKERQLQQQQQQRYLQQQQLSATNALIPHVQAQAQPPISSP 867
Query: 377 TGQKS------ISLTGSQPDATASGATTPGGSSSQ 405
S S S AT S TP S Q
Sbjct: 868 QQNSSQAQPQNSSQQVSVSPATPSSPLTPMSSQHQ 972
>TC78816 similar to GP|14190377|gb|AAK55669.1 AT4g00850/A_TM018A10_22
{Arabidopsis thaliana}, partial (43%)
Length = 1150
Score = 43.1 bits (100), Expect = 3e-04
Identities = 41/134 (30%), Positives = 55/134 (40%), Gaps = 8/134 (5%)
Frame = +1
Query: 285 PASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQK-SMGQPQLHQQQPSPQQ 343
PA P ++ H QQ +Q QQ F Q+ G P HQ Q Q
Sbjct: 424 PALPPQMAPHPAMQQGF----YMQHPQAAAMAQQQGMFPQKMPMQFGNP--HQMQDQQHQ 585
Query: 344 QQQQQLLQPQQQSQL---QASVHQQQH-LHSPRVAGPTGQKSIS---LTGSQPDATASGA 396
QQQQQL Q Q Q+ ++ H +H+ G +G + GS+ DA+ +G
Sbjct: 586 QQQQQLHQQAMQGQMGLRPGGINNGMHPMHNEAALGGSGSGGPNDGRGGGSKQDASEAGT 765
Query: 397 TTPGGSSSQGTEAA 410
G QGT AA
Sbjct: 766 ---AGGDGQGTSAA 798
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.309 0.121 0.328
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,204,371
Number of Sequences: 36976
Number of extensions: 196727
Number of successful extensions: 3390
Number of sequences better than 10.0: 190
Number of HSP's better than 10.0 without gapping: 1926
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2429
length of query: 570
length of database: 9,014,727
effective HSP length: 101
effective length of query: 469
effective length of database: 5,280,151
effective search space: 2476390819
effective search space used: 2476390819
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144645.6


