

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144645.5 - phase: 0
(254 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
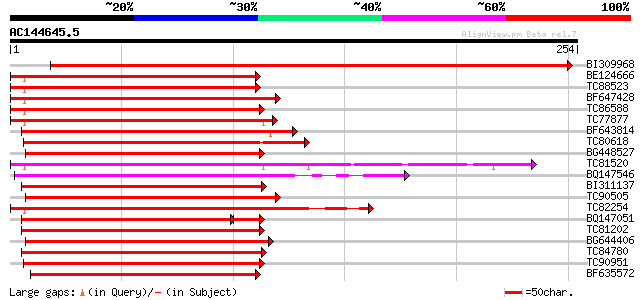
Score E
Sequences producing significant alignments: (bits) Value
BI309968 similar to GP|13346178|gb| BNLGHi233 {Gossypium hirsutu... 483 e-137
BE124666 similar to PIR|JQ0957|JQ0 myb-related protein 330 - gar... 159 8e-40
TC88523 similar to PIR|T05690|T05690 myb-related transcription f... 158 2e-39
BF647428 homologue to PIR|T51621|T51 myb-like protein [imported]... 151 3e-37
TC86588 similar to GP|5139814|dbj|BAA81736.1 GmMYB29B2 {Glycine ... 150 5e-37
TC77877 similar to GP|5139806|dbj|BAA81732.1 GmMYB29A2 {Glycine ... 150 5e-37
BF643814 similar to GP|19386839|db putative myb-related protein ... 150 6e-37
TC80618 homologue to PIR|T06455|T06455 Myb26 protein - garden pe... 148 2e-36
BG448527 similar to PIR|T51509|T51 probable transcription factor... 148 2e-36
TC81520 similar to PIR|D86470|D86470 hypothetical protein AAD460... 148 2e-36
BQ147546 similar to PIR|H96706|H96 probable transcription factor... 147 4e-36
BI311137 similar to GP|17380966|gb putative transcription factor... 147 5e-36
TC90505 similar to GP|3941528|gb|AAC83640.1| putative transcript... 145 1e-35
TC82254 similar to GP|22266673|dbj|BAC07543. myb-related transcr... 145 1e-35
BQ147051 similar to GP|23476311|gb myb-like transcription factor... 142 2e-35
TC81202 homologue to GP|17380966|gb|AAL36295.1 putative transcri... 144 4e-35
BG644406 homologue to PIR|T07398|T07 myb-related transcription f... 143 6e-35
TC84780 homologue to PIR|F85021|F85021 probable transcription fa... 143 7e-35
TC90951 homologue to PIR|T06455|T06455 Myb26 protein - garden pe... 142 9e-35
BF635572 similar to GP|7673084|gb| An2 protein {Petunia x hybrid... 140 4e-34
>BI309968 similar to GP|13346178|gb| BNLGHi233 {Gossypium hirsutum}, partial
(40%)
Length = 759
Score = 483 bits (1242), Expect = e-137
Identities = 234/234 (100%), Positives = 234/234 (100%)
Frame = +3
Query: 19 DILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHISTDEEDMIIRLH 78
DILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHISTDEEDMIIRLH
Sbjct: 21 DILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHISTDEEDMIIRLH 200
Query: 79 RLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSSLPSPSSVSLRHNHGKCG 138
RLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSSLPSPSSVSLRHNHGKCG
Sbjct: 201 RLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSSLPSPSSVSLRHNHGKCG 380
Query: 139 HVAPEAPKPRRLKAVHQYKILEKNSGSEYDQGSDETSIADFFIDFDHQDQLMVGDDESNS 198
HVAPEAPKPRRLKAVHQYKILEKNSGSEYDQGSDETSIADFFIDFDHQDQLMVGDDESNS
Sbjct: 381 HVAPEAPKPRRLKAVHQYKILEKNSGSEYDQGSDETSIADFFIDFDHQDQLMVGDDESNS 560
Query: 199 KIPQMEDHKVSSTNSTHSSSSPSDHCHLLAEKFDPQEILLDVELKKMASFLGLE 252
KIPQMEDHKVSSTNSTHSSSSPSDHCHLLAEKFDPQEILLDVELKKMASFLGLE
Sbjct: 561 KIPQMEDHKVSSTNSTHSSSSPSDHCHLLAEKFDPQEILLDVELKKMASFLGLE 722
>BE124666 similar to PIR|JQ0957|JQ0 myb-related protein 330 - garden
snapdragon, partial (48%)
Length = 536
Score = 159 bits (403), Expect = 8e-40
Identities = 71/116 (61%), Positives = 91/116 (78%), Gaps = 4/116 (3%)
Frame = +3
Query: 1 MVRSP----KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNY 56
M RSP + NKGAW++EED+ L Y+ +HGEG W+ + + AGL RCGKSCR RW+NY
Sbjct: 27 MGRSPCCEKEHTNKGAWTKEEDERLINYIKLHGEGCWRSLPKAAGLLRCGKSCRLRWINY 206
Query: 57 LKPGIKRGHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKL 112
L+P +KRG+ + E+D+II LH LLGN+WSLIA RLPGRTDNEIKNYWNT++ +KL
Sbjct: 207 LRPDLKRGNFTEQEDDLIINLHSLLGNKWSLIAARLPGRTDNEIKNYWNTHIKRKL 374
>TC88523 similar to PIR|T05690|T05690 myb-related transcription factor MYB4
- Arabidopsis thaliana, partial (54%)
Length = 1292
Score = 158 bits (399), Expect = 2e-39
Identities = 71/116 (61%), Positives = 91/116 (78%), Gaps = 4/116 (3%)
Frame = +1
Query: 1 MVRSP----KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNY 56
M RSP NKGAW++EEDD L Y+ HGEG W+ + + AGL RCGKSCR RW+NY
Sbjct: 271 MGRSPCCEKAHTNKGAWTKEEDDRLISYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWINY 450
Query: 57 LKPGIKRGHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKL 112
L+P +KRG+ + +E+++II+LH LLGN+WSLIA RLPGRTDNEIKNYWNT++ +KL
Sbjct: 451 LRPDLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRKL 618
>BF647428 homologue to PIR|T51621|T51 myb-like protein [imported] -
Arabidopsis thaliana, partial (44%)
Length = 689
Score = 151 bits (381), Expect = 3e-37
Identities = 75/125 (60%), Positives = 86/125 (68%), Gaps = 4/125 (3%)
Frame = +2
Query: 1 MVRSP----KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNY 56
MVR P + V KG W+ EED IL Y+ HG G W+ V N GL RC KSCR RW NY
Sbjct: 92 MVRPPCCDKEGVKKGPWTPEEDIILVSYIQEHGPGNWKAVPTNTGLLRCSKSCRLRWTNY 271
Query: 57 LKPGIKRGHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQP 116
L+PGIKRG+ + EE MII L LLGNRW+ IA LP RTDN+IKNYWNT L KKL+K
Sbjct: 272 LRPGIKRGNFTDQEEKMIIHLQALLGNRWAAIAAYLPQRTDNDIKNYWNTYLKKKLKKLE 451
Query: 117 TSSSS 121
TS+SS
Sbjct: 452 TSTSS 466
>TC86588 similar to GP|5139814|dbj|BAA81736.1 GmMYB29B2 {Glycine max},
partial (85%)
Length = 1389
Score = 150 bits (379), Expect = 5e-37
Identities = 68/118 (57%), Positives = 89/118 (74%), Gaps = 4/118 (3%)
Frame = +2
Query: 1 MVRSP----KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNY 56
MVR+P + +G WS EED+IL+ Y+ HG G W+ + + AGL RCGKSCR RW+NY
Sbjct: 107 MVRAPCCEKMGLKRGPWSLEEDEILTSYIQKHGHGNWRALPKLAGLLRCGKSCRLRWINY 286
Query: 57 LKPGIKRGHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQK 114
L+P IKRG+ + +EE+ II+LH +LGNRWS IA +LPGRTDNEIKN W+T+L KKL +
Sbjct: 287 LRPDIKRGNFTNEEEENIIKLHEMLGNRWSAIAAKLPGRTDNEIKNVWHTHLKKKLNQ 460
>TC77877 similar to GP|5139806|dbj|BAA81732.1 GmMYB29A2 {Glycine max},
partial (78%)
Length = 1156
Score = 150 bits (379), Expect = 5e-37
Identities = 70/128 (54%), Positives = 93/128 (71%), Gaps = 8/128 (6%)
Frame = +3
Query: 1 MVRSP----KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNY 56
MVR+P K + KG W+ EED+IL+ Y+ HG W+ + ++AGL RCGKSCR RW+NY
Sbjct: 66 MVRAPCCEKKGLKKGPWTLEEDEILTSYINKHGHSNWRALPKHAGLLRCGKSCRLRWINY 245
Query: 57 LKPGIKRGHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKL---- 112
LKP IKRG+ + +EE+ II++H LGNRWS IA +LPGRTDNEIKN W+T+L K+L
Sbjct: 246 LKPDIKRGNFTNEEEETIIKMHESLGNRWSAIAAKLPGRTDNEIKNVWHTHLKKRLLNTN 425
Query: 113 QKQPTSSS 120
QP S++
Sbjct: 426 NNQPNSNT 449
>BF643814 similar to GP|19386839|db putative myb-related protein {Oryza
sativa (japonica cultivar-group)}, partial (49%)
Length = 545
Score = 150 bits (378), Expect = 6e-37
Identities = 66/127 (51%), Positives = 94/127 (73%), Gaps = 3/127 (2%)
Frame = +1
Query: 6 KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGH 65
+ NKGAWS+EED+ L Y+ GEG W+ + + AGL RCGKSCR RW+NYL+P +KRG+
Sbjct: 16 QHTNKGAWSKEEDERLINYIKQQGEGCWRSLPKAAGLARCGKSCRLRWINYLRPDLKRGN 195
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQ---PTSSSSL 122
+ +E+++II LH ++GN+WS IA++LPGRTDNEIKNYWNT++ +KL + PT+ L
Sbjct: 196 FTHEEDELIISLHAMVGNKWSQIAQKLPGRTDNEIKNYWNTHIKRKLYSRGIDPTTHQPL 375
Query: 123 PSPSSVS 129
+ S +
Sbjct: 376 KTLSGTA 396
>TC80618 homologue to PIR|T06455|T06455 Myb26 protein - garden pea, partial
(57%)
Length = 799
Score = 148 bits (374), Expect = 2e-36
Identities = 69/128 (53%), Positives = 90/128 (69%)
Frame = +2
Query: 7 EVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHI 66
+V KG W+ EED IL Y+ HGEG W +A++AGLKR GKSCR RWLNYL+P ++RG+I
Sbjct: 86 DVRKGPWTMEEDLILINYIANHGEGVWNSLAKSAGLKRTGKSCRLRWLNYLRPDVRRGNI 265
Query: 67 STDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSSLPSPS 126
+ +E+ +II LH GNRWS IAK LPGRTDNEIKNYW T + K + KQ +S S S
Sbjct: 266 TPEEQLLIIELHAKWGNRWSKIAKHLPGRTDNEIKNYWRTRIQKHI-KQAENSQSQEQGS 442
Query: 127 SVSLRHNH 134
+ + ++
Sbjct: 443 DIQINDDN 466
>BG448527 similar to PIR|T51509|T51 probable transcription factor (MYB9) -
Arabidopsis thaliana, partial (40%)
Length = 633
Score = 148 bits (373), Expect = 2e-36
Identities = 66/107 (61%), Positives = 80/107 (74%)
Frame = +1
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
V KG W++EED+ L Y+ HG G W +++ AGL RCGKSCR RW NYL+P IKRG +
Sbjct: 73 VKKGPWTQEEDEKLIDYINKHGHGNWGTLSKRAGLNRCGKSCRLRWTNYLRPDIKRGKFT 252
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQK 114
+EE +II LH +LGN+WS IA LPGRTDNEIKNYWNTN+ KKL K
Sbjct: 253 DEEERVIINLHSVLGNKWSKIAAHLPGRTDNEIKNYWNTNIRKKLLK 393
>TC81520 similar to PIR|D86470|D86470 hypothetical protein AAD46010.1
[imported] - Arabidopsis thaliana, partial (39%)
Length = 1259
Score = 148 bits (373), Expect = 2e-36
Identities = 96/266 (36%), Positives = 133/266 (49%), Gaps = 30/266 (11%)
Frame = +2
Query: 1 MVRSP----KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNY 56
M RSP + KG W+ EED L +++ HG G W+ + + AGL RCGKSCR RW NY
Sbjct: 56 MGRSPCCDESGLKKGPWTPEEDQKLVEHIQQHGHGSWRALPKLAGLNRCGKSCRLRWTNY 235
Query: 57 LKPGIKRGHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKL---- 112
L+P IKRG S +EE I+ LH +LGN+WS IA LPGRTDNEIKN+WNT+L KKL
Sbjct: 236 LRPDIKRGKFSQEEEQTILHLHSILGNKWSAIATHLPGRTDNEIKNFWNTHLKKKLIQMG 415
Query: 113 ----QKQPTSSSSLPSPSSVSLRH----------------NHGKCGHVAPEAPKPRRLKA 152
QP + P ++L + H + EA +L+
Sbjct: 416 FDPMTHQPRTDLVSTIPYLLALANMTEIIDHQNQSSWDDQQHAAMSSLQAEAVHLAKLQC 595
Query: 153 VHQYKILEKNSGSEYDQGSDETSIADFFIDFDHQDQLMVGDDESNSKIPQMEDHKVSSTN 212
+ QY + NS + S + + + + + L + + SN K + D S +
Sbjct: 596 L-QYLLQSSNSSININNNSYDQNA--MITNMEQEQPLSLLNTISNVKENTIMD--CSQLD 760
Query: 213 STH--SSSSPSDHCHLLAEKFDPQEI 236
STH S S P H +L DPQ++
Sbjct: 761 STHAASFSQPLHHQSVLPHFLDPQQV 838
>BQ147546 similar to PIR|H96706|H96 probable transcription factor T22E19.5
[imported] - Arabidopsis thaliana, partial (48%)
Length = 690
Score = 147 bits (371), Expect = 4e-36
Identities = 76/177 (42%), Positives = 107/177 (59%)
Frame = +3
Query: 3 RSPKEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIK 62
+ E+ +G W+ EED +L Y+ HGEG+W +A++AGLKR GKSCR RWLNYLKP IK
Sbjct: 147 KEENELRRGPWTLEEDSLLIHYIARHGEGRWNMLAKSAGLKRTGKSCRLRWLNYLKPDIK 326
Query: 63 RGHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSSL 122
RG+++ E+ +I+ LH GNRWS IA+ LPGRTDNEIKNYW T + K+ ++ S S
Sbjct: 327 RGNLTPQEQLLILELHSKXGNRWSKIAQHLPGRTDNEIKNYWRTRVQKQARQLNIESGSK 506
Query: 123 PSPSSVSLRHNHGKCGHVAPEAPKPRRLKAVHQYKILEKNSGSEYDQGSDETSIADF 179
+V KC + PR L+ +E+N+ +D S T++ +F
Sbjct: 507 RFIDAV-------KCFWM------PRLLQK------MEQNTSPSFDNSSSMTNMNNF 620
>BI311137 similar to GP|17380966|gb putative transcription factor
{Arabidopsis thaliana}, partial (35%)
Length = 761
Score = 147 bits (370), Expect = 5e-36
Identities = 66/110 (60%), Positives = 83/110 (75%)
Frame = +1
Query: 6 KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGH 65
+++ KG WS EED+ L Y+ HG G W V + AGL+RCGKSCR RW+NYL+P +KRG
Sbjct: 175 QKLRKGLWSPEEDEKLLNYITKHGHGCWSSVPKLAGLQRCGKSCRLRWINYLRPDLKRGA 354
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQ 115
S EE+ II LH LLGNRWS IA +LPGRTDNEIKN WN++L KKL+++
Sbjct: 355 FSEQEENSIIELHALLGNRWSQIAAQLPGRTDNEIKNLWNSSLKKKLKQK 504
>TC90505 similar to GP|3941528|gb|AAC83640.1| putative transcription factor
{Arabidopsis thaliana}, partial (49%)
Length = 984
Score = 145 bits (367), Expect = 1e-35
Identities = 67/114 (58%), Positives = 80/114 (69%)
Frame = +1
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
V KG W+ EED IL Y+ HG G W+ V GL RC KSCR RW NYL+PGIKRG+ +
Sbjct: 190 VKKGPWTPEEDIILVTYIQEHGPGNWRAVPTKTGLSRCSKSCRLRWTNYLRPGIKRGNFT 369
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSS 121
EE MII L LLGNRW+ IA LP RTDN+IKNYWNT+L KKL+K T +++
Sbjct: 370 EQEEKMIIHLQDLLGNRWAAIASYLPQRTDNDIKNYWNTHLKKKLKKSQTGNNA 531
>TC82254 similar to GP|22266673|dbj|BAC07543. myb-related transcription
factor VlMYBB1-1 {Vitis labrusca x Vitis vinifera},
partial (46%)
Length = 754
Score = 145 bits (366), Expect = 1e-35
Identities = 77/167 (46%), Positives = 103/167 (61%), Gaps = 4/167 (2%)
Frame = +2
Query: 1 MVRSP----KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNY 56
MVR+P + KG W+ EED L YV +G W+++ + AGL+RCGKSCR RWLNY
Sbjct: 98 MVRTPYCDKNGLRKGTWTPEEDKKLIAYVTRYGCWNWRQLPKFAGLERCGKSCRLRWLNY 277
Query: 57 LKPGIKRGHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQP 116
L+P IKRG S +EE+ II+LH LGNRW++I+ LPGRTDNEIKNYW+T + K L K
Sbjct: 278 LRPDIKRGSFSHEEEETIIKLHEKLGNRWTMISANLPGRTDNEIKNYWHTTIKKTLVKNK 457
Query: 117 TSSSSLPSPSSVSLRHNHGKCGHVAPEAPKPRRLKAVHQYKILEKNS 163
+++ + S NH P KP++L ++LE NS
Sbjct: 458 SNTKRGTKKAKDSNSKNH-------PTMEKPKKLG-----EMLENNS 562
>BQ147051 similar to GP|23476311|gb myb-like transcription factor 6
{Gossypium hirsutum}, partial (53%)
Length = 445
Score = 142 bits (358), Expect(2) = 2e-35
Identities = 62/96 (64%), Positives = 77/96 (79%)
Frame = +3
Query: 6 KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGH 65
+ +NKGAWS++ED L Y+ +HGEG W + + AGL RCGKSCR RWLNYL+P IKRG
Sbjct: 84 ENINKGAWSKQEDQKLIDYIQVHGEGCWGSIPKAAGLHRCGKSCRLRWLNYLRPDIKRGI 263
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIK 101
+ DEED+II+LH LLGNRW+LIA R+PGRTDNE+K
Sbjct: 264 FAQDEEDLIIKLHALLGNRWALIAGRVPGRTDNEVK 371
Score = 23.9 bits (50), Expect(2) = 2e-35
Identities = 8/14 (57%), Positives = 12/14 (85%)
Frame = +2
Query: 101 KNYWNTNLSKKLQK 114
+NYWN+++ KKL K
Sbjct: 368 ENYWNSHIRKKLIK 409
>TC81202 homologue to GP|17380966|gb|AAL36295.1 putative transcription
factor {Arabidopsis thaliana}, partial (40%)
Length = 1506
Score = 144 bits (362), Expect = 4e-35
Identities = 64/109 (58%), Positives = 82/109 (74%)
Frame = +2
Query: 6 KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGH 65
+++ KG WS EED+ L Y+ HG G W V + AGL+RCGKSCR RW+NYL+P +KRG
Sbjct: 86 QKLRKGLWSPEEDEKLLNYITKHGHGCWSSVPKLAGLQRCGKSCRLRWINYLRPDLKRGA 265
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQK 114
S EE++II LH +LGNRWS IA +LPGRTDNEIKN WN+ L K+L++
Sbjct: 266 FSQQEENLIIELHAVLGNRWSQIAAQLPGRTDNEIKNLWNSCLKKRLRQ 412
>BG644406 homologue to PIR|T07398|T07 myb-related transcription factor THM6 -
tomato, partial (42%)
Length = 762
Score = 143 bits (361), Expect = 6e-35
Identities = 66/111 (59%), Positives = 80/111 (71%)
Frame = +1
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
V KG W+ EED +L +V HG G W+ V + GL+RC KSCR RW NYL+PGIKRG +
Sbjct: 178 VKKGPWTPEEDIMLVSFVQEHGPGNWRTVPTHTGLRRCSKSCRLRWTNYLRPGIKRGSFT 357
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTS 118
EE MII+L LLGN+W+ IA LP RTDN+IKNYWNT+L KKL+K TS
Sbjct: 358 DQEEKMIIQLQALLGNKWAAIASYLPERTDNDIKNYWNTHLKKKLKKLETS 510
>TC84780 homologue to PIR|F85021|F85021 probable transcription factor
[imported] - Arabidopsis thaliana, partial (39%)
Length = 971
Score = 143 bits (360), Expect = 7e-35
Identities = 63/110 (57%), Positives = 84/110 (76%)
Frame = +1
Query: 6 KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGH 65
+++ KG WS EED+ L ++ +G G W V + AGL+RCGKSCR RW+NYL+P +KRG
Sbjct: 130 QKLRKGLWSPEEDEKLLNHITKYGHGCWSSVPKQAGLQRCGKSCRLRWINYLRPDLKRGT 309
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQ 115
S +EE++II LH +LGNRWS IA +LPGRTDNEIKN WN+ L KKL+++
Sbjct: 310 FSQEEENLIIELHSVLGNRWSQIAAQLPGRTDNEIKNLWNSCLKKKLRQK 459
>TC90951 homologue to PIR|T06455|T06455 Myb26 protein - garden pea, partial
(83%)
Length = 661
Score = 142 bits (359), Expect = 9e-35
Identities = 63/108 (58%), Positives = 81/108 (74%)
Frame = +3
Query: 7 EVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHI 66
EV KG W+ EED IL Y+ HGEG W +A+ AGLKR GKSCR RWLNYL+P ++RG+I
Sbjct: 99 EVRKGPWTMEEDLILINYIANHGEGVWNSLAKAAGLKRTGKSCRLRWLNYLRPDVRRGNI 278
Query: 67 STDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQK 114
+ +E+ +I+ LH GNRWS IAK LPGRTDNEIKN+W T + K +++
Sbjct: 279 TPEEQLLIMELHAKWGNRWSKIAKHLPGRTDNEIKNFWRTRIQKHIKQ 422
>BF635572 similar to GP|7673084|gb| An2 protein {Petunia x hybrida}, partial
(40%)
Length = 367
Score = 140 bits (354), Expect = 4e-34
Identities = 63/103 (61%), Positives = 77/103 (74%)
Frame = +2
Query: 10 KGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHISTD 69
KGAW+ EED++L + +GEGKW V + GL RC KSCR RWLNYL P I R + D
Sbjct: 2 KGAWTYEEDNLLKACIHKYGEGKWHLVPKRTGLNRCRKSCRLRWLNYLNPAINRESFAED 181
Query: 70 EEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKL 112
E DMI+RLH+LLGN+WSLIA RLPGRT N++ NYW+TNL KK+
Sbjct: 182 EIDMILRLHKLLGNKWSLIAARLPGRTANDVXNYWHTNLRKKV 310
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.131 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,870,309
Number of Sequences: 36976
Number of extensions: 140267
Number of successful extensions: 902
Number of sequences better than 10.0: 136
Number of HSP's better than 10.0 without gapping: 869
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 881
length of query: 254
length of database: 9,014,727
effective HSP length: 94
effective length of query: 160
effective length of database: 5,538,983
effective search space: 886237280
effective search space used: 886237280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC144645.5


