

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144645.3 + phase: 0 /pseudo
(993 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
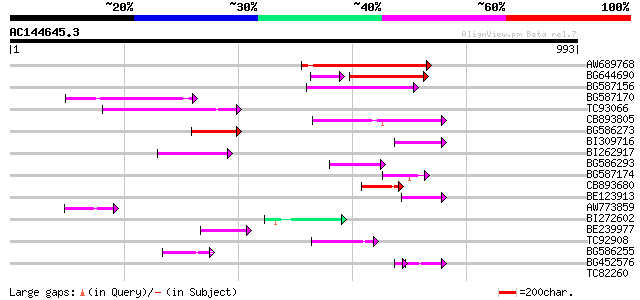
Score E
Sequences producing significant alignments: (bits) Value
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 208 9e-54
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 128 1e-38
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 156 3e-38
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 130 3e-30
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 127 2e-29
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 125 8e-29
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 93 6e-19
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 80 4e-15
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 79 1e-14
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 74 2e-13
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 65 1e-10
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 65 1e-10
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 60 3e-09
AW773859 58 2e-08
BI272602 GP|11602812|db avenaIII {Gallus gallus}, partial (3%) 57 3e-08
BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryz... 54 4e-07
TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol po... 50 6e-06
BG586255 similar to GP|7682800|gb Hypothetical protein T15F17.l ... 49 1e-05
BG452576 weakly similar to GP|12005223|gb|A reverse transcriptas... 41 3e-05
TC82260 similar to GP|19310478|gb|AAL84973.1 AT3g05350/T12H1_32 ... 40 0.004
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 208 bits (529), Expect = 9e-54
Identities = 109/230 (47%), Positives = 140/230 (60%), Gaps = 1/230 (0%)
Frame = +1
Query: 511 THSMHGIHKPKIQFNLTTSITSSPLPHN-PKAALSDSNWKAAMLDEFDALIKNKTWELVP 569
T + G KPK+ + P P + LSD W AM E+ ALI NKTW+LVP
Sbjct: 4 TRTKTGKLKPKV-------FLTEPCPQDCENLPLSDPRWLQAMKTEYKALIDNKTWDLVP 162
Query: 570 RPQNVNFIRSMWIFRHKKKSDGSFERYKARLVGDGRSQ*VGVDCDETFSPVVKPTTIRIV 629
P + I W++R K+ DGS ++KARLV G SQ +G D ETFSPV+KP TIR++
Sbjct: 163 LPPHKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFSQTLGCDYTETFSPVIKPVTIRLI 342
Query: 630 LTIALSKSWPIHQLDVKNVFLHGELQETVYMHQPMGFRDPIHPDYVCLLKKSLYGLKQAP 689
LTIA++ W I Q+D+ N FL+G LQE VYM QP GF + + VC L KSLYGLKQAP
Sbjct: 343 LTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGF-EAANKSLVCKLNKSLYGLKQAP 519
Query: 690 RAWYQRFADFAFTIGFSHSKSDHSLFIYRKGNDMTYILLYVDDIILTASS 739
RAWY+ GF+ S+ D SL IY + Y+ +YVDDI++T SS
Sbjct: 520 RAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNGACIYLXIYVDDILITGSS 669
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 128 bits (322), Expect(2) = 1e-38
Identities = 62/139 (44%), Positives = 93/139 (66%)
Frame = -2
Query: 595 RYKARLVGDGRSQ*VGVDCDETFSPVVKPTTIRIVLTIALSKSWPIHQLDVKNVFLHGEL 654
R K++LV G +Q G+D DE FSPV + IRI++ A + ++Q+DVK+ F++G+L
Sbjct: 418 RNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFINGDL 239
Query: 655 QETVYMHQPMGFRDPIHPDYVCLLKKSLYGLKQAPRAWYQRFADFAFTIGFSHSKSDHSL 714
+E V++ QP GF D P++V L K+LYGLKQAPRAWY+R + F GF K D++L
Sbjct: 238 KEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDNTL 59
Query: 715 FIYRKGNDMTYILLYVDDI 733
F+ ++ ++ I +YVDDI
Sbjct: 58 FLLKRE*ELLIIQVYVDDI 2
Score = 50.8 bits (120), Expect(2) = 1e-38
Identities = 23/59 (38%), Positives = 35/59 (58%)
Frame = -3
Query: 528 TSITSSPLPHNPKAALSDSNWKAAMLDEFDALIKNKTWELVPRPQNVNFIRSMWIFRHK 586
++ SS P N K AL D++W +M +E ++K W LVPRP+ I + W+FR+K
Sbjct: 609 SAFISSIEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVFRNK 433
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 156 bits (395), Expect = 3e-38
Identities = 82/196 (41%), Positives = 115/196 (57%)
Frame = -1
Query: 520 PKIQFNLTTSITSSPLPHNPKAALSDSNWKAAMLDEFDALIKNKTWELVPRPQNVNFIRS 579
P+ ++ + +P + + A+ D WK ++ E A+IKN TW P+ + S
Sbjct: 606 PEAHCAFMVNLNENHIPRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSS 427
Query: 580 MWIFRHKKKSDGSFERYKARLVGDGRSQ*VGVDCDETFSPVVKPTTIRIVLTIALSKSWP 639
WIF K K+DGS ER K RLV G + G D ETF+PV K TIRIVL++A++ W
Sbjct: 426 RWIFTIKYKADGSIERKKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWG 247
Query: 640 IHQLDVKNVFLHGELQETVYMHQPMGFRDPIHPDYVCLLKKSLYGLKQAPRAWYQRFADF 699
+ Q+DVKN FL GEL++ VYM+ P G + V LKK++YGLKQ+PRAWY + +
Sbjct: 246 LWQMDVKNAFLQGELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTT 67
Query: 700 AFTIGFSHSKSDHSLF 715
GF S+ DH+LF
Sbjct: 66 LNGRGFRKSELDHTLF 19
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 130 bits (326), Expect = 3e-30
Identities = 71/234 (30%), Positives = 120/234 (50%), Gaps = 3/234 (1%)
Frame = -3
Query: 99 GDLY---PLTTTTSHQSTSPATFAALSPTLWHNRLGHPGANVLSFLNKNKFIECKQISSP 155
GDLY L ++++ + ++ + LWH RLGHP L+ + E K
Sbjct: 671 GDLYMLEKLDPVSNYKCSFTSSSSLNKDALWHARLGHPHGRALNLMLPGVVFENKN---- 504
Query: 156 SICQSCIYGKHVKLPFSISNSTTSKPFDIIHSDLWTSPILSSAGHKYYLFFLDDYTNFVW 215
C++CI GKH K F +++ FD+I++DLWT+P LS HKY++ F+D+ + + W
Sbjct: 503 --CEACILGKHCKNVFPRTSTVYENCFDLIYTDLWTAPSLSRDNHKYFVTFIDEKSKYTW 330
Query: 216 TFPIGRKSQVPSIFSSFHAFIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSC 275
I K +V F +F A++ + IK + DNG E+ + F +G++ + SC
Sbjct: 329 LTLIPSKDRVIDAFKNFQAYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHGILHQTSC 150
Query: 276 PHTSPQNGKAERKIRAINNFIRTSLAHSSMPPSFWHHALQITTYLQNILPSKIL 329
P+T QNG A+RK + + R+ + ++ + YL N +P+K+L
Sbjct: 149 PYTPQQNGVAKRKNKHLMEVARSLMFQAN---------VSTACYLINWIPTKVL 15
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 127 bits (319), Expect = 2e-29
Identities = 79/249 (31%), Positives = 116/249 (45%), Gaps = 5/249 (2%)
Frame = +1
Query: 163 YGKHVKLPFSISNSTTSKPFDIIHSDLW-TSPILSSAGHKYYLFFLDDYTNFVWTFPIGR 221
+G K+ FS + T D IHSDLW S + S G +Y + +DD+ VW + +
Sbjct: 40 FGNRKKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRY 219
Query: 222 KSQVPSIFSSFHAFIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSCPHTSPQ 281
K++ F + ++TQ G +K DN EF + F +FC +G+ + P Q
Sbjct: 220 KNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQ 399
Query: 282 NGKAERKIRAINNFIRTSLAHSSM--PPSFWHHALQITTYLQNILPSKILSHHSPTQYLY 339
NG AER IR + R L+++ + W A +L N P L P
Sbjct: 400 NGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWS 579
Query: 340 HRDPSYTHLRVFGCLCYPLFPSTAINKLQARSTPCAFLGYPQNHRGYK--CYDLSSKKII 397
Y++LR+FGC Y L KL R+ C FL Y +GY+ C D S+K+I
Sbjct: 580 GNLVDYSNLRIFGCPAYALVND---GKLAPRAGECIFLSYASESKGYRLWCSDPKSQKLI 750
Query: 398 ISRHVIFDE 406
+SR V F+E
Sbjct: 751 LSRDVTFNE 777
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 125 bits (314), Expect = 8e-29
Identities = 80/243 (32%), Positives = 118/243 (47%), Gaps = 7/243 (2%)
Frame = +3
Query: 530 ITSSPLPHNPKAALSDSNWKAAMLDEFDALIKNKTWELVPRPQNVNFIRSMWIFRHKKKS 589
+T + P + A+ W+A+M +E +A +N TWEL I WIF+ K
Sbjct: 24 LTMTSDPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNE 203
Query: 590 DGSFERYKARLVGDGRSQ*VGVDCDETFSPVVKPTTIRIVLTIALSKSWPIHQLDVKNVF 649
+G E+YKARLV G SQ GVD E F+PV + TIR+V+ +A Q+ V
Sbjct: 204 NGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAA-------QIKRDGVC 362
Query: 650 L------HGELQETVYMHQPMGFRDPIHPDYVCLLKKSLYGLKQAPRAWYQRFADFAFTI 703
+ H ++ + + R +K++LYGLKQAPRAWY R +
Sbjct: 363 IS*M*KAHSCMEN*MRKFLLINHRVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTKE 542
Query: 704 GFSHSKSDHSLFI-YRKGNDMTYILLYVDDIILTASSDVLRRSIMSLLASEFAMKDLGTL 762
GF +H+LF+ +G + I LYVDD+I + + + + EF M DLG +
Sbjct: 543 GFEKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKM 722
Query: 763 SYF 765
YF
Sbjct: 723 HYF 731
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 92.8 bits (229), Expect = 6e-19
Identities = 41/88 (46%), Positives = 58/88 (65%)
Frame = -2
Query: 319 YLQNILPSKILSHHSPTQYLYHRDPSYTHLRVFGCLCYPLFPSTAINKLQARSTPCAFLG 378
YL N +P+++L +P + L R PS T++RVFGCLCY L P NKL+ARS F+G
Sbjct: 698 YLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMFIG 519
Query: 379 YPQNHRGYKCYDLSSKKIIISRHVIFDE 406
Y +GYKCYD ++++++SR V F E
Sbjct: 518 YSTTQKGYKCYDPEARRVLVSRDVKFIE 435
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 80.1 bits (196), Expect = 4e-15
Identities = 39/91 (42%), Positives = 55/91 (59%)
Frame = +2
Query: 675 VCLLKKSLYGLKQAPRAWYQRFADFAFTIGFSHSKSDHSLFIYRKGNDMTYILLYVDDII 734
VC L+KS+YGLKQA R WY + ++ + G+ S SD SLF K + T +L+YVDDI+
Sbjct: 20 VCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDIV 199
Query: 735 LTASSDVLRRSIMSLLASEFAMKDLGTLSYF 765
L + + + L F +KDLG+L YF
Sbjct: 200 LAGNDISEIQHVKCFLIDRFKIKDLGSLRYF 292
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 78.6 bits (192), Expect = 1e-14
Identities = 44/131 (33%), Positives = 62/131 (46%)
Frame = +1
Query: 260 FTQFCHKNGMVFRFSCPHTSPQNGKAERKIRAINNFIRTSLAHSSMPPSFWHHALQITTY 319
F F +K + HT QNG AER R + R L + M SFW A++ Y
Sbjct: 31 F*HFVNKRVL*GNSXVAHTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACY 210
Query: 320 LQNILPSKILSHHSPTQYLYHRDPSYTHLRVFGCLCYPLFPSTAINKLQARSTPCAFLGY 379
+ N PS ++ +P + + Y+ L VFGC Y ++ S KL +S C FLGY
Sbjct: 211 VINRSPSTVIDLKTPMEMWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGY 390
Query: 380 PQNHRGYKCYD 390
N +GY +D
Sbjct: 391 ADNVKGYXLWD 423
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 74.3 bits (181), Expect = 2e-13
Identities = 39/97 (40%), Positives = 58/97 (59%)
Frame = +2
Query: 561 KNKTWELVPRPQNVNFIRSMWIFRHKKKSDGSFERYKARLVGDGRSQ*VGVDCDETFSPV 620
+ +T +LV +P V I WI++ K+ DG+ +YKARLV G + G+D DE F+PV
Sbjct: 47 QKQTLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPV 226
Query: 621 VKPTTIRIVLTIALSKSWPIHQLDVKNVFLHGELQET 657
V+ TI ++L +A + IH +DVK FL+G T
Sbjct: 227 VRIETI*LLLALAATNGC*IHHIDVKIAFLNGHFVGT 337
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse
transcriptase homolog - rape retrotransposon copia-like
(fragment), partial (84%)
Length = 249
Score = 65.5 bits (158), Expect = 1e-10
Identities = 37/91 (40%), Positives = 53/91 (57%), Gaps = 9/91 (9%)
Frame = -1
Query: 653 ELQETVYMHQPMGFRDPIHPDYVCLLKKSLYGLKQAPRAWYQRFADF---------AFTI 703
EL+E +YM QP GF P D+VC L+KSLYGLKQ+PR WY+RF + T+
Sbjct: 249 ELEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRFDSYRSSWATTGVLMTV 70
Query: 704 GFSHSKSDHSLFIYRKGNDMTYILLYVDDII 734
+ ++ S +I Y++LYVDD++
Sbjct: 69 VST*TR*RMSRYI--------YLVLYVDDML 1
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 65.1 bits (157), Expect = 1e-10
Identities = 36/74 (48%), Positives = 46/74 (61%)
Frame = -2
Query: 617 FSPVVKPTTIRIVLTIALSKSWPIHQLDVKNVFLHGELQETVYMHQPMGFRDPIHPDYVC 676
F P+VK TI +L+I ++ + LDVK FL G+L E +YMHQP GF + V
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*EV-GKMVG 378
Query: 677 LLKKSLYGLKQAPR 690
LKKS+YGLKQ PR
Sbjct: 377 KLKKSMYGLKQGPR 336
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 60.5 bits (145), Expect = 3e-09
Identities = 32/80 (40%), Positives = 44/80 (55%), Gaps = 1/80 (1%)
Frame = +1
Query: 687 QAPRAWYQRFADFAFTIGFSHSKSDHSLFIYRKGNDMTYILL-YVDDIILTASSDVLRRS 745
Q+PR W+ RF G+ ++DH++FI IL+ YVDDI LT +
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 746 IMSLLASEFAMKDLGTLSYF 765
+ +LLA EF +KDLG L YF
Sbjct: 181 LKNLLAEEFEIKDLGNLKYF 240
>AW773859
Length = 538
Score = 57.8 bits (138), Expect = 2e-08
Identities = 33/95 (34%), Positives = 44/95 (45%), Gaps = 1/95 (1%)
Frame = -3
Query: 97 STGDLYPLTTTTSHQSTSPATFAALSPTLWHNRLGHPGANVLSFLNKN-KFIECKQISSP 155
+T D T +S P LWH RLGH L L+ N FI I
Sbjct: 314 TTQDTPSATIASSQVQPQPQPTFLPQEALWHFRLGHLSNRKLLSLHSNFPFIT---IDQN 144
Query: 156 SICQSCIYGKHVKLPFSISNSTTSKPFDIIHSDLW 190
S+C C Y +H KLPF +S + SK +++ H D+W
Sbjct: 143 SVCDICHYSRHKKLPFQLSTNRASKCYELFHFDIW 39
>BI272602 GP|11602812|db avenaIII {Gallus gallus}, partial (3%)
Length = 488
Score = 57.0 bits (136), Expect = 3e-08
Identities = 44/149 (29%), Positives = 58/149 (38%), Gaps = 5/149 (3%)
Frame = +1
Query: 446 PEPRKIESPQPATTPASPI-----NVTNQSILPPSPMSINQLPHPLVSTELTSPTHTPQQ 500
P ++ ++PASP N+T PP P PT P
Sbjct: 34 PNSNEVAQNIQISSPASPTSNELANITTSPTPPPPP----------------PPTARPHN 165
Query: 501 IHQEPPRTIATHSMHGIHKPKIQFNLTTSITSSPLPHNPKAALSDSNWKAAMLDEFDALI 560
H I T S + I K + NL P N AL D +W++ M DEFDAL
Sbjct: 166 NHTTNVGGIITRSQNNIVKTIKKLNLHVRPLCPIEPSNITQALRDHDWRSIMQDEFDALQ 345
Query: 561 KNKTWELVPRPQNVNFIRSMWIFRHKKKS 589
N TW+LV R N + F ++ KS
Sbjct: 346 NNNTWDLVSRSFAQNLVGCKLGFSYQTKS 432
>BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 514
Score = 53.5 bits (127), Expect = 4e-07
Identities = 28/90 (31%), Positives = 43/90 (47%)
Frame = -2
Query: 334 PTQYLYHRDPSYTHLRVFGCLCYPLFPSTAINKLQARSTPCAFLGYPQNHRGYKCYDLSS 393
P+ Y Y + R+FGC + S +K R+ C F+ Y +GY+CY S
Sbjct: 501 PSFYPYVPTSNNLTPRIFGCTSFVHIHSDGRSKFDHRALKCVFIRYSSTQKGYRCYHPPS 322
Query: 394 KKIIISRHVIFDETQFPFAKTHTSSTHTYE 423
+K +SR V F E + F + +SS Y+
Sbjct: 321 RKYFVSRDVTFHEQESYFGQN*SSSGGKYK 232
>TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol polyprotein
{Oryza sativa}, partial (1%)
Length = 638
Score = 49.7 bits (117), Expect = 6e-06
Identities = 39/118 (33%), Positives = 56/118 (47%), Gaps = 1/118 (0%)
Frame = +2
Query: 529 SITSSPLPHNPKAALSDSNWKAAMLDEFDALIKNKTWELVPRPQNVNFIRSMWIFRHKKK 588
+ITS+ P N A W AAM E L +N T L P + ++ +++ K
Sbjct: 260 AITSNIEPKNYVQAAQ*QEWLAAMEQEIQVLEENNTSTLEPLREGKKWVDCRPVYKIIHK 439
Query: 589 SDGSFERYKARLVGDGRSQ*VGVDC-DETFSPVVKPTTIRIVLTIALSKSWPIHQLDV 645
++G E+YKA+LV Q G D D S K R +LTIA +K +H +DV
Sbjct: 440 ANGEIEKYKAQLVAKDFVQVEGEDF*DLCLSN--KDDNCRCLLTIAAAKG*QLHLMDV 607
>BG586255 similar to GP|7682800|gb Hypothetical protein T15F17.l {Arabidopsis
thaliana}, partial (12%)
Length = 436
Score = 48.9 bits (115), Expect = 1e-05
Identities = 33/92 (35%), Positives = 43/92 (45%)
Frame = -1
Query: 268 GMVFRFSCPHTSPQNGKAERKIRAINNFIRTSLAHSSMPPSFWHHALQITTYLQNILPSK 327
G V+ S H QNG AE I+ I R L S P S W HA+ L I PS
Sbjct: 415 G*VWNTSVVHVHTQNGLAESFIKRIQLITRPLLMRSKQPVSAWGHAVLHAAELIRIRPSS 236
Query: 328 ILSHHSPTQYLYHRDPSYTHLRVFGCLCYPLF 359
+SP+Q L P +H++ FG YP++
Sbjct: 235 -EHKYSPSQLLSGHVPDVSHIKTFG---YPVY 152
>BG452576 weakly similar to GP|12005223|gb|A reverse transcriptase-like
protein {Spiranthes hongkongensis}, partial (25%)
Length = 676
Score = 40.8 bits (94), Expect(2) = 3e-05
Identities = 22/72 (30%), Positives = 35/72 (48%)
Frame = +2
Query: 693 YQRFADFAFTIGFSHSKSDHSLFIYRKGNDMTYILLYVDDIILTASSDVLRRSIMSLLAS 752
Y + ++G+ S +DHS+F + G T L+Y+DDI+ + + S L
Sbjct: 245 Y*KLLSTLISLGYKQSPNDHSIFSF--GRRFTIFLVYLDDIVFARDDHSETQLVKSHLDK 418
Query: 753 EFAMKDLGTLSY 764
F + LGTL Y
Sbjct: 419 NFIIIGLGTLHY 454
Score = 25.8 bits (55), Expect(2) = 3e-05
Identities = 11/22 (50%), Positives = 13/22 (59%)
Frame = +3
Query: 675 VCLLKKSLYGLKQAPRAWYQRF 696
VC S+YGLK A R Y +F
Sbjct: 165 VCKFHNSIYGLKHAHRQ*YSKF 230
>TC82260 similar to GP|19310478|gb|AAL84973.1 AT3g05350/T12H1_32
{Arabidopsis thaliana}, partial (20%)
Length = 700
Score = 40.0 bits (92), Expect = 0.004
Identities = 34/106 (32%), Positives = 47/106 (44%), Gaps = 9/106 (8%)
Frame = +3
Query: 434 HYHLQ---NDPKQDEPEPRKIESPQPATTPASPINVTNQSILP------PSPMSINQLPH 484
H+HLQ N P Q P S Q ++T ++PIN+TN P P P N LP+
Sbjct: 18 HHHLQL*RNSPSQSTPPSPLSISIQSSSTNSNPINITNPLSSPSAIAPLPIPSQRNHLPN 197
Query: 485 PLVSTELTSPTHTPQQIHQEPPRTIATHSMHGIHKPKIQFNLTTSI 530
V T PT TP PP +++ + P LT+S+
Sbjct: 198 S-VKTVPQIPTPTP----NSPPSAVSSPN------PMSPLTLTSSL 302
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.332 0.142 0.470
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,859,718
Number of Sequences: 36976
Number of extensions: 762728
Number of successful extensions: 6485
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 4681
Number of HSP's successfully gapped in prelim test: 227
Number of HSP's that attempted gapping in prelim test: 1340
Number of HSP's gapped (non-prelim): 5338
length of query: 993
length of database: 9,014,727
effective HSP length: 106
effective length of query: 887
effective length of database: 5,095,271
effective search space: 4519505377
effective search space used: 4519505377
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144645.3


