

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144564.13 - phase: 0
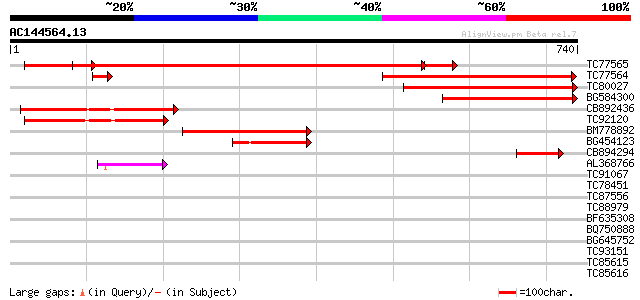
(740 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77565 similar to GP|21592665|gb|AAM64614.1 Isp4-like protein {... 829 0.0
TC77564 similar to GP|20466754|gb|AAM20694.1 Isp4-like protein {... 486 e-137
TC80027 similar to GP|15451020|gb|AAK96781.1 Unknown protein {Ar... 311 6e-85
BG584300 similar to GP|6983868|dbj EST D25093(R3139) corresponds... 258 6e-69
CB892436 similar to PIR|T08925|T08 isp4 protein homolog T15N24.4... 236 2e-62
TC92120 similar to PIR|T01900|T01900 isp4 protein homolog T12H20... 221 1e-57
BM778892 similar to GP|6983868|dbj EST D25093(R3139) corresponds... 192 5e-49
BG454123 similar to PIR|T01900|T01 isp4 protein homolog T12H20.7... 122 5e-28
CB894294 similar to GP|20160527|dbj putative sexual differentiat... 105 6e-23
AL368766 GP|20466754|gb Isp4-like protein {Arabidopsis thaliana}... 75 1e-13
TC91067 similar to PIR|H96681|H96681 protein F1E22.10 [imported]... 41 0.002
TC78451 similar to GP|9279625|dbj|BAB01083.1 gb|AAF23830.1~gene_... 35 0.10
TC87556 similar to GP|14517530|gb|AAK62655.1 At1g48370/F11A17_27... 35 0.14
TC88979 similar to PIR|H96601|H96601 hypothetical protein T6H22.... 31 2.0
BF635308 similar to GP|10177061|db beta-galactosidase {Arabidops... 30 2.6
BQ750888 GP|13992574|em glutenin HMW subunit 1Ax {Triticum timop... 30 4.4
BG645752 similar to GP|21672287|gb| hv711N16.16 {Hordeum vulgare... 29 5.7
TC93151 homologue to GP|22136928|gb|AAM91808.1 unknown protein {... 29 5.7
TC85615 similar to GP|10178248|dbj|BAB11680. glucosyltransferase... 29 5.7
TC85616 similar to GP|10178248|dbj|BAB11680. glucosyltransferase... 29 5.7
>TC77565 similar to GP|21592665|gb|AAM64614.1 Isp4-like protein {Arabidopsis
thaliana}, partial (71%)
Length = 1698
Score = 829 bits (2141), Expect(2) = 0.0
Identities = 395/462 (85%), Positives = 427/462 (91%)
Frame = +1
Query: 82 TLPLGHLMASVLPSKTFRIPGFGSKRFSFNPGPFNMKEHVLITIFANAGAAFGSGSSYAV 141
TLP+GH MA++LP+ FRIPGFG+K+ S NPGPFNMKEHVLITIFANAG+AFGSGS YAV
Sbjct: 190 TLPIGHFMAAILPTTKFRIPGFGTKKLSLNPGPFNMKEHVLITIFANAGSAFGSGSPYAV 369
Query: 142 GIVNIIKAFYGRNISFLAAWLLIITTQVLGYGWAGLLRKYVVEPAHMWWPGTLVQVSLFR 201
GIVNIIKAFYGR+ISF AAWLLIITTQVLGYGWAGLLRKYVVEPAHMWWP TLVQVSLFR
Sbjct: 370 GIVNIIKAFYGRSISFHAAWLLIITTQVLGYGWAGLLRKYVVEPAHMWWPSTLVQVSLFR 549
Query: 202 TLHEKDDNPHQFSRAKFFFIALVCSFTWYIVPGYLFTTLTSISWVCWVFSKSVTAQQIGS 261
LHEKDD+ H+ SRAKFFFIALVCSF+WY++PGYLFTTLTSISWVCWVF+KSVTAQQ+GS
Sbjct: 550 ALHEKDDD-HRISRAKFFFIALVCSFSWYVIPGYLFTTLTSISWVCWVFTKSVTAQQLGS 726
Query: 262 GMNGLGLGALTLDWSAVASFLFSPLISPFFAIVNVFVGYALLVYAVIPIAYWGFNVYGAN 321
GM GLG+GA+TLDW+AVASFLFSPLISPFFAIVNVFVGY L++YAV+PIAYWG N Y A
Sbjct: 727 GMKGLGIGAITLDWTAVASFLFSPLISPFFAIVNVFVGYVLIIYAVVPIAYWGLNAYNAK 906
Query: 322 RFSIFSSDLYTAQGQPYNISNIVNDKFEIDLAKYHEQGRIHLSTFFALSYGFGFATIAST 381
F +FSS L+TAQGQ YNIS IVNDKFE+D AKY EQGRIHLS FF+L+YGFGFATIAST
Sbjct: 907 TFPLFSSHLFTAQGQKYNISAIVNDKFELDEAKYAEQGRIHLSVFFSLTYGFGFATIAST 1086
Query: 382 VTHVACFYGREIMERYRASKNGKEDIHTKLMKNYKDIPSWWFYLLLGVTFVVSLMICIFL 441
+THVACFYGREIMERYRAS GKEDIHTKLMK YKDIPSWWFY LL VT VSL++CIFL
Sbjct: 1087LTHVACFYGREIMERYRASSKGKEDIHTKLMKRYKDIPSWWFYALLVVTLAVSLVLCIFL 1266
Query: 442 NDQIQMPWWGLLIASALAFIFTLPISIITATTNQTPGLNIITEYIFGIIYPGRPIANVCF 501
NDQIQMPWWGLL A ALAF FTLPISIITATTNQTPGLNIITEY+FG+IYPGRPIANVCF
Sbjct: 1267NDQIQMPWWGLLFAGALAFAFTLPISIITATTNQTPGLNIITEYVFGLIYPGRPIANVCF 1446
Query: 502 KTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLA 543
KTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGT+LA
Sbjct: 1447KTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTMLA 1572
Score = 119 bits (297), Expect = 5e-27
Identities = 61/94 (64%), Positives = 66/94 (69%)
Frame = +3
Query: 20 EESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFAYRTEPLIITQITVQ 79
E SPIEEVRLTV TDDPT PVWTFRMWFLGL+SC+LLSFLNQFFAYRTEPLIIT ITVQ
Sbjct: 3 EISPIEEVRLTVTNTDDPTQPVWTFRMWFLGLISCSLLSFLNQFFAYRTEPLIITLITVQ 182
Query: 80 VATLPLGHLMASVLPSKTFRIPGFGSKRFSFNPG 113
VA P L + + P K+ PG
Sbjct: 183 VAYTPYRSLHGGYSANHEVQNPRVRDKKVVIEPG 284
Score = 80.5 bits (197), Expect(2) = 0.0
Identities = 34/43 (79%), Positives = 38/43 (88%)
Frame = +2
Query: 542 LAGTINIGVAWWLLDSVKNICNKDLLPKGSPWTCPSDRVFFDA 584
L GTINIGVAWWLL+SV+NIC+ DLLP+GSPWTCP D FFDA
Sbjct: 1568 LPGTINIGVAWWLLNSVENICHDDLLPEGSPWTCPGDPCFFDA 1696
>TC77564 similar to GP|20466754|gb|AAM20694.1 Isp4-like protein {Arabidopsis
thaliana}, partial (37%)
Length = 1275
Score = 486 bits (1251), Expect = e-137
Identities = 221/253 (87%), Positives = 237/253 (93%)
Frame = +1
Query: 487 FGIIYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTI 546
FG+IYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGT+LAGTI
Sbjct: 76 FGLIYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTMLAGTI 255
Query: 547 NIGVAWWLLDSVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTL 606
NIGVAWWLL+SV+NIC+ DLLP+GSPWTCP DRVFFDASV+WGLVGPKRIFGS G YS +
Sbjct: 256 NIGVAWWLLNSVENICHDDLLPEGSPWTCPGDRVFFDASVIWGLVGPKRIFGSKGNYSAM 435
Query: 607 NWFFLGGAIGPILVWLLHKAFPKQSWIPLINLPVLLGATGMMPPATALNYNSWIIVGTIF 666
NWFF+GGAIGPI+VWLLHKAFP+QSWI LINLPVLLGAT MMPPAT LNYNSWI VGTIF
Sbjct: 436 NWFFIGGAIGPIIVWLLHKAFPRQSWIALINLPVLLGATAMMPPATPLNYNSWIFVGTIF 615
Query: 667 NFFIFRYRKKWWQRYNYVLSAALDAGVAFMAVLLYLALGLENVSLNWWGTAGEHCPLAAC 726
NFF+FRYRKKWWQRYNYVLSAALD GVAFM VLLY +L LEN S++WWGT GEHCPLA+C
Sbjct: 616 NFFVFRYRKKWWQRYNYVLSAALDTGVAFMTVLLYFSLSLENRSISWWGTEGEHCPLASC 795
Query: 727 PTAKGIVVDGCPV 739
PTAKGI VDGCPV
Sbjct: 796 PTAKGIAVDGCPV 834
Score = 54.3 bits (129), Expect = 2e-07
Identities = 24/26 (92%), Positives = 25/26 (95%)
Frame = +1
Query: 109 SFNPGPFNMKEHVLITIFANAGAAFG 134
S NPGPFNMKEHVLITIFANAG+AFG
Sbjct: 4 SLNPGPFNMKEHVLITIFANAGSAFG 81
>TC80027 similar to GP|15451020|gb|AAK96781.1 Unknown protein {Arabidopsis
thaliana}, partial (30%)
Length = 926
Score = 311 bits (797), Expect = 6e-85
Identities = 136/227 (59%), Positives = 168/227 (73%)
Frame = +1
Query: 514 SFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAWWLLDSVKNICNKDLLPKGSPW 573
+FL + KLGHYMKIPPR M+ Q +GT++AG +N+ VAWW+LDS+K+IC D SPW
Sbjct: 1 AFLQNLKLGHYMKIPPRCMYTAQLVGTLVAGVVNLSVAWWMLDSIKDICMDDKAHHDSPW 180
Query: 574 TCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNWFFLGGAIGPILVWLLHKAFPKQSWI 633
TCP RV FDASV+WGL+GPKR+FG G Y L W FL GA+ P+ VW+ K +P + WI
Sbjct: 181 TCPKYRVTFDASVIWGLIGPKRLFGPGGLYRNLVWLFLVGAVLPVPVWVFSKIYPNKKWI 360
Query: 634 PLINLPVLLGATGMMPPATALNYNSWIIVGTIFNFFIFRYRKKWWQRYNYVLSAALDAGV 693
PLIN+PV+ MPPAT N SW++ G IFN+F+FR+ K+WWQ+YNYVLSAALDAG
Sbjct: 361 PLINIPVISYGFAGMPPATPTNIASWLLTGMIFNYFVFRFHKRWWQKYNYVLSAALDAGT 540
Query: 694 AFMAVLLYLALGLENVSLNWWGTAGEHCPLAACPTAKGIVVDGCPVF 740
AFM VL++ AL +L WWGT +HCPLA CPTA GIVVDGCPVF
Sbjct: 541 AFMGVLIFFALQNAGHTLKWWGTELDHCPLATCPTAPGIVVDGCPVF 681
>BG584300 similar to GP|6983868|dbj EST D25093(R3139) corresponds to a region
of the predicted gene.~Similar to Arabidopsis thaliana,
partial (21%)
Length = 755
Score = 258 bits (659), Expect = 6e-69
Identities = 107/175 (61%), Positives = 138/175 (78%)
Frame = +3
Query: 566 LLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNWFFLGGAIGPILVWLLHK 625
+LP GSPWTCP D VF+D+SVVWGL+GP+RIFG+LG YS +NWFFL GA+ P +VWL HK
Sbjct: 27 VLPAGSPWTCPGDHVFYDSSVVWGLIGPRRIFGNLGHYSAINWFFLVGAVAPFIVWLAHK 206
Query: 626 AFPKQSWIPLINLPVLLGATGMMPPATALNYNSWIIVGTIFNFFIFRYRKKWWQRYNYVL 685
A P + WI LI +PV+LGA MPPAT +NY SW++VG F +RY + WW R+NYVL
Sbjct: 207 ALPDKQWIKLITMPVILGALTEMPPATPVNYTSWVLVGFASGFVAYRYYRGWWTRHNYVL 386
Query: 686 SAALDAGVAFMAVLLYLALGLENVSLNWWGTAGEHCPLAACPTAKGIVVDGCPVF 740
S ALDAG+AFM VL+YL LG++++SL+WWG+ + CPLA+CPTA G++ GCP+F
Sbjct: 387 SGALDAGLAFMGVLIYLCLGMQHISLDWWGSDPDRCPLASCPTAPGVISAGCPLF 551
>CB892436 similar to PIR|T08925|T08 isp4 protein homolog T15N24.40 -
Arabidopsis thaliana, partial (24%)
Length = 707
Score = 236 bits (603), Expect = 2e-62
Identities = 113/207 (54%), Positives = 152/207 (72%), Gaps = 1/207 (0%)
Frame = +2
Query: 15 EEEEEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFAYRTEPLIIT 74
++ E ++SPIE+VRLTV TDDPT P TFR W +GL C +L+F+NQFF YRT PL IT
Sbjct: 95 DDTEVDDSPIEQVRLTVSTTDDPTQPALTFRTWIIGLACCIVLAFVNQFFGYRTNPLTIT 274
Query: 75 QITVQVATLPLGHLMASVLPSKTFRIPGFGSKRFSFNPGPFNMKEHVLITIFANAGAAFG 134
++ Q+ +LP+G LMA+ LP+ +++P F F+ NPGPFN+KEH LITIFA+AGA
Sbjct: 275 AVSAQIVSLPIGKLMAATLPTTIYKVP-FTKWSFTLNPGPFNLKEHALITIFASAGA--- 442
Query: 135 SGSSYAVGIVNIIKAFYGRNISFLAAWLLIITTQVLGYGWAGLLRKYVVEPAHMWWPGTL 194
G YA+ I+ I+KAFY RNI+ +AA+LL ITTQ+LGYGWAG+ R+++V+ +MWWP L
Sbjct: 443 -GGVYAINIITIVKAFYHRNINPIAAFLLAITTQMLGYGWAGMFRRFLVDSPYMWWPFNL 619
Query: 195 VQVSLFRTLHEKDDN-PHQFSRAKFFF 220
VQVSLFR HEK+ + +R +F F
Sbjct: 620 VQVSLFRAFHEKEKRLKGRTTRLQFLF 700
>TC92120 similar to PIR|T01900|T01900 isp4 protein homolog T12H20.7 -
Arabidopsis thaliana, partial (25%)
Length = 647
Score = 221 bits (562), Expect = 1e-57
Identities = 111/189 (58%), Positives = 139/189 (72%), Gaps = 1/189 (0%)
Frame = +2
Query: 20 EESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFAYRTEPLIITQITVQ 79
E SPIE+V LTV TDDP+LPV+TFR W LG L+C LLSFLNQFF +R EPL +T I+ Q
Sbjct: 62 ENSPIEQVALTVPVTDDPSLPVFTFRTWTLGTLACVLLSFLNQFFGFRREPLSVTAISAQ 241
Query: 80 VATLPLGHLMASVLPSKTFRIPGFGSK-RFSFNPGPFNMKEHVLITIFANAGAAFGSGSS 138
+A +PLGHLMAS + + F G K F+ NPG FN+KEHVLITIFA++GAA S
Sbjct: 242 IAVVPLGHLMASTITKRVFM---KGKKWEFTLNPGKFNVKEHVLITIFASSGAA----SV 400
Query: 139 YAVGIVNIIKAFYGRNISFLAAWLLIITTQVLGYGWAGLLRKYVVEPAHMWWPGTLVQVS 198
YA+ V+ +K FY + I+ L A L+++TTQVLG+GWAG+ +Y+VEPA MWWP LVQVS
Sbjct: 401 YAIHFVSTVKVFYRKEITVLVALLVVLTTQVLGFGWAGVFXRYLVEPAGMWWPQNLVQVS 580
Query: 199 LFRTLHEKD 207
LFR L K+
Sbjct: 581 LFRALXXKE 607
>BM778892 similar to GP|6983868|dbj EST D25093(R3139) corresponds to a region
of the predicted gene.~Similar to Arabidopsis thaliana,
partial (22%)
Length = 852
Score = 192 bits (487), Expect = 5e-49
Identities = 84/168 (50%), Positives = 119/168 (70%)
Frame = +1
Query: 226 SFTWYIVPGYLFTTLTSISWVCWVFSKSVTAQQIGSGMNGLGLGALTLDWSAVASFLFSP 285
SF +Y++PGYLF LTS+SW+CWVF S+ AQQ+GSG++GLG+GA+ DWS++ S+L SP
Sbjct: 1 SFAYYVLPGYLFPMLTSLSWICWVFPNSIIAQQLGSGLHGLGVGAIGFDWSSICSYLGSP 180
Query: 286 LISPFFAIVNVFVGYALLVYAVIPIAYWGFNVYGANRFSIFSSDLYTAQGQPYNISNIVN 345
L SP+FA N+ G+ + +Y V+PIAY G N+Y F IFS L+ GQ YNIS I++
Sbjct: 181 LASPWFATANIAAGFGIFMYVVVPIAY-GLNLYHGRSFPIFSDGLFNTNGQEYNISAIID 357
Query: 346 DKFEIDLAKYHEQGRIHLSTFFALSYGFGFATIASTVTHVACFYGREI 393
F +DL Y +G ++LST FA+SYG FA +++ + HV F+GR +
Sbjct: 358 SNFHLDLDAYQREGPLYLSTMFAMSYGIDFACLSAILVHVLLFHGRYV 501
>BG454123 similar to PIR|T01900|T01 isp4 protein homolog T12H20.7 -
Arabidopsis thaliana, partial (13%)
Length = 654
Score = 122 bits (306), Expect = 5e-28
Identities = 53/104 (50%), Positives = 77/104 (73%)
Frame = +1
Query: 291 FAIVNVFVGYALLVYAVIPIAYWGFNVYGANRFSIFSSDLYTAQGQPYNISNIVNDKFEI 350
FA NV VG+ ++Y + P+ YW FNVY A F IFS+ L+T++G+ YNI+ I++ F +
Sbjct: 1 FATANVAVGFVFVMYILTPLCYW-FNVYNAKTFPIFSNQLFTSKGEIYNITEIIDSNFHM 177
Query: 351 DLAKYHEQGRIHLSTFFALSYGFGFATIASTVTHVACFYGREIM 394
DLA Y +QGR+HLSTFFA++YG GFA + +T+ HVA F+GR ++
Sbjct: 178 DLAAYEKQGRLHLSTFFAMTYGVGFAALTATIVHVALFHGRYVL 309
>CB894294 similar to GP|20160527|dbj putative sexual differentiation process
protein Isp4 {Oryza sativa (japonica cultivar-group)},
partial (8%)
Length = 190
Score = 105 bits (262), Expect = 6e-23
Identities = 44/62 (70%), Positives = 53/62 (84%)
Frame = +2
Query: 662 VGTIFNFFIFRYRKKWWQRYNYVLSAALDAGVAFMAVLLYLALGLENVSLNWWGTAGEHC 721
VGTIF+FF+FRYR++WWQRY+YVLSAALD GVAFM VLLY +L LE ++WWG G+HC
Sbjct: 5 VGTIFSFFVFRYRRRWWQRYSYVLSAALDTGVAFMTVLLYFSLSLELRGISWWGADGDHC 184
Query: 722 PL 723
PL
Sbjct: 185 PL 190
>AL368766 GP|20466754|gb Isp4-like protein {Arabidopsis thaliana}, partial
(4%)
Length = 516
Score = 74.7 bits (182), Expect = 1e-13
Identities = 47/99 (47%), Positives = 56/99 (56%), Gaps = 7/99 (7%)
Frame = -1
Query: 115 FNMKEHVLI-------TIFANAGAAFGSGSSYAVGIVNIIKAFYGRNISFLAAWLLIITT 167
F +K+HVLI T F N + A+ V I + G A + +
Sbjct: 297 FILKKHVLIFFKKFKNTKFFNLFHNMP*KHTLAIAFVIIEQNCDGNGPLIYAFFSDNVYL 118
Query: 168 QVLGYGWAGLLRKYVVEPAHMWWPGTLVQVSLFRTLHEK 206
VLGYGWAGLLRKYVVEPAHMWWP TLVQVSLFR ++K
Sbjct: 117 *VLGYGWAGLLRKYVVEPAHMWWPSTLVQVSLFRYDNKK 1
>TC91067 similar to PIR|H96681|H96681 protein F1E22.10 [imported] -
Arabidopsis thaliana, partial (46%)
Length = 1223
Score = 40.8 bits (94), Expect = 0.002
Identities = 44/180 (24%), Positives = 76/180 (41%), Gaps = 8/180 (4%)
Frame = +3
Query: 418 IPSWWF---YLLLGVTFVVSLMICIFLNDQIQMPWWGLL----IASALAFIFTLPISIIT 470
IPSW+ Y+++G+ ++++ Q+ W+ ++ IA ALAF +
Sbjct: 258 IPSWFSIAGYVIIGIISIITIPHIFH-----QIKWYHVILIYIIAPALAFCNAYGCGLTD 422
Query: 471 ATTNQTPG-LNIITEYIFGIIYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPP 529
+ T G L I T + G +A + ++ A + DFK G+ P
Sbjct: 423 WSLASTYGKLAIFTIGAWAGSANGGVLAGLAACGVMMNIVSTASDLMQDFKTGYMTLASP 602
Query: 530 RSMFLVQFIGTVLAGTINIGVAWWLLDSVKNICNKDLLPKGSPWTCPSDRVFFDASVVWG 589
+SMF+ Q IGT + I+ V W + L GS + P VF + +++ G
Sbjct: 603 KSMFVSQVIGTAMGCIISPCVFWLFYHAFGT-----LGQPGSAYPAPYALVFRNIAIIGG 767
>TC78451 similar to GP|9279625|dbj|BAB01083.1
gb|AAF23830.1~gene_id:MOJ10.9~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (88%)
Length = 2452
Score = 35.0 bits (79), Expect = 0.10
Identities = 52/202 (25%), Positives = 86/202 (41%), Gaps = 10/202 (4%)
Frame = +2
Query: 453 LIASALAFIFTLPISIIT---ATTNQTPGLNIITEYIFGIIYPGRPIANVCFKTYGYISM 509
++A ALAF + + A+T GL II + G IA V +
Sbjct: 1448 ILAPALAFCNSYGTGLTDWSLASTYGKIGLFIIAA---AVGTNGGVIAGVASCAVMMSIV 1618
Query: 510 AQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAWWLLDSVKNICNKDLLPK 569
A A + DFK G+ +SMF+ Q IGT + G I + +W+ + +I + D
Sbjct: 1619 ATAADLMQDFKTGYLTLSSAKSMFVSQLIGTAM-GCIIAPLTFWMFWTAFDIGSPD---- 1783
Query: 570 GSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEY--STLNWFFLGGAIGPILVWLLHKAF 627
P+ P +F + +++ G+ G F L +Y FF + + LL
Sbjct: 1784 -GPYKAPYAVIFREMAIL-GVEG----FSELPKYCMEMCGGFFAAA----LAINLLRDVT 1933
Query: 628 PKQ--SWIPL---INLPVLLGA 644
PK+ +IP+ + +P +GA
Sbjct: 1934 PKKYSQYIPIPMAMAVPFYIGA 1999
>TC87556 similar to GP|14517530|gb|AAK62655.1 At1g48370/F11A17_27
{Arabidopsis thaliana}, partial (48%)
Length = 1309
Score = 34.7 bits (78), Expect = 0.14
Identities = 17/44 (38%), Positives = 24/44 (53%)
Frame = +1
Query: 509 MAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAW 552
+A A + DFK G+ PRS+F+ Q IGT + I+ V W
Sbjct: 517 VATASDLMQDFKTGYLTLASPRSVFVSQIIGTTMGCIISPCVFW 648
>TC88979 similar to PIR|H96601|H96601 hypothetical protein T6H22.14
[imported] - Arabidopsis thaliana, partial (89%)
Length = 1616
Score = 30.8 bits (68), Expect = 2.0
Identities = 15/38 (39%), Positives = 23/38 (60%)
Frame = -1
Query: 555 LDSVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVG 592
++S IC+K+LLP+ SP TC + AS ++ L G
Sbjct: 500 MESTSRICDKNLLPRPSP*TC----TLYKASNIYKLYG 399
>BF635308 similar to GP|10177061|db beta-galactosidase {Arabidopsis
thaliana}, partial (16%)
Length = 507
Score = 30.4 bits (67), Expect = 2.6
Identities = 29/123 (23%), Positives = 48/123 (38%), Gaps = 12/123 (9%)
Frame = +2
Query: 416 KDIPSWWFYLLLGVTFVVSLMICIFLNDQIQMPWWGLLIASALAFIFTLPISIITATTNQ 475
K + WW +LL + VSL C + L+I +F+ I +T
Sbjct: 71 KTMGEWWRFLLHALILTVSL--CTVHGANVTYDRTSLVINGHHKILFSGSIHYPRSTPQM 244
Query: 476 TP---------GLNIITEYIFGIIY---PGRPIANVCFKTYGYISMAQAVSFLSDFKLGH 523
P GL++I Y+F ++ G+ N F G+I QA ++G
Sbjct: 245 WPDLISKAKEGGLDVIQTYVFWNLHEPQQGQYEFNGRFDLXGFIKEIQAQGLYVTLRIGP 424
Query: 524 YMK 526
Y++
Sbjct: 425 YIE 433
>BQ750888 GP|13992574|em glutenin HMW subunit 1Ax {Triticum timopheevii},
partial (1%)
Length = 721
Score = 29.6 bits (65), Expect = 4.4
Identities = 18/61 (29%), Positives = 28/61 (45%)
Frame = +2
Query: 512 AVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAWWLLDSVKNICNKDLLPKGS 571
A + +SDF++G ++ PP F Q IG ++ + GV L + I P G
Sbjct: 479 ATALVSDFRVGFLLRTPPHLQFYAQAIGGAVSIFLAPGVVVLFLSAYPCIYQ----PSGD 646
Query: 572 P 572
P
Sbjct: 647 P 649
>BG645752 similar to GP|21672287|gb| hv711N16.16 {Hordeum vulgare}, partial
(20%)
Length = 650
Score = 29.3 bits (64), Expect = 5.7
Identities = 19/72 (26%), Positives = 34/72 (46%)
Frame = +1
Query: 285 PLISPFFAIVNVFVGYALLVYAVIPIAYWGFNVYGANRFSIFSSDLYTAQGQPYNISNIV 344
P++ F A + +FV ALL ++V+ W ++ S++ + +ISNI+
Sbjct: 367 PVVEAFRARLKIFVPIALLAFSVMVSVIW-------TNDTLERSNVVYSSIDKLSISNIL 525
Query: 345 NDKFEIDLAKYH 356
I L +YH
Sbjct: 526 FGSNRISLKQYH 561
>TC93151 homologue to GP|22136928|gb|AAM91808.1 unknown protein {Arabidopsis
thaliana}, partial (4%)
Length = 657
Score = 29.3 bits (64), Expect = 5.7
Identities = 22/66 (33%), Positives = 30/66 (45%), Gaps = 3/66 (4%)
Frame = +3
Query: 76 ITVQVATL---PLGHLMASVLPSKTFRIPGFGSKRFSFNPGPFNMKEHVLITIFANAGAA 132
IT +AT+ P G ++ S T FGS F +P PF+ + T A +
Sbjct: 93 ITFSLATMSGFPFGSSSSTQSSSSTSAF-SFGSSPFGSSPSPFSTTTNPTSTATAPSSFN 269
Query: 133 FGSGSS 138
FGS SS
Sbjct: 270 FGSSSS 287
>TC85615 similar to GP|10178248|dbj|BAB11680. glucosyltransferase-like protein
{Arabidopsis thaliana}, partial (90%)
Length = 2067
Score = 29.3 bits (64), Expect = 5.7
Identities = 16/48 (33%), Positives = 25/48 (51%), Gaps = 1/48 (2%)
Frame = -1
Query: 381 TVTHVACFYGREIMERYRASKNGK-EDIHTKLMKNYKDIPSWWFYLLL 427
TV V F R I++ G+ I T+ ++N+KD S WFY ++
Sbjct: 1962 TVDLVQRFVARRILDSLLVLCRGETRTISTRHLRNFKDFGSKWFYTMV 1819
>TC85616 similar to GP|10178248|dbj|BAB11680. glucosyltransferase-like protein
{Arabidopsis thaliana}, partial (17%)
Length = 1175
Score = 29.3 bits (64), Expect = 5.7
Identities = 16/48 (33%), Positives = 25/48 (51%), Gaps = 1/48 (2%)
Frame = -2
Query: 381 TVTHVACFYGREIMERYRASKNGK-EDIHTKLMKNYKDIPSWWFYLLL 427
TV V F R I++ G+ I T+ ++N+KD S WFY ++
Sbjct: 1072 TVDLVQRFVARRILDSLLVLCRGETRTISTRHLRNFKDFGSKWFYTMV 929
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.327 0.142 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,952,039
Number of Sequences: 36976
Number of extensions: 444627
Number of successful extensions: 3535
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 2570
Number of HSP's successfully gapped in prelim test: 122
Number of HSP's that attempted gapping in prelim test: 777
Number of HSP's gapped (non-prelim): 2841
length of query: 740
length of database: 9,014,727
effective HSP length: 103
effective length of query: 637
effective length of database: 5,206,199
effective search space: 3316348763
effective search space used: 3316348763
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144564.13


