

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144563.4 + phase: 0
(588 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
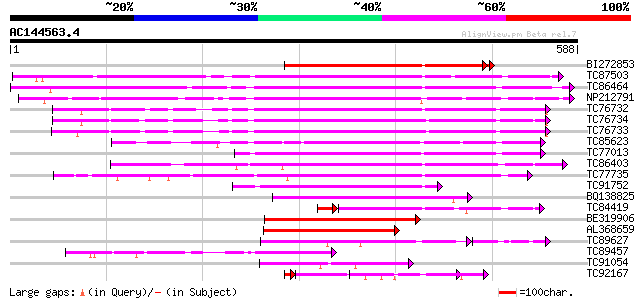
Score E
Sequences producing significant alignments: (bits) Value
BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-... 332 7e-92
TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precurs... 318 3e-87
TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Cit... 312 2e-85
NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1 295 3e-80
TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.... 295 3e-80
TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 279 2e-75
TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 277 7e-75
TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 ... 259 2e-69
TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylestera... 246 1e-65
TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF028... 238 4e-63
TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectineste... 230 1e-60
TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectineste... 170 2e-42
BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like pr... 154 1e-37
TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor ... 147 6e-37
BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precurso... 149 4e-36
AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886... 127 1e-29
TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pec... 102 2e-26
TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE... 110 1e-24
TC91054 similar to GP|20521423|dbj|BAB91933. putative pectin met... 89 6e-18
TC92167 similar to PIR|T05203|T05203 pectinesterase homolog F4I1... 76 3e-14
>BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-like protein
- maize, partial (25%)
Length = 678
Score = 332 bits (850), Expect(2) = 7e-92
Identities = 163/210 (77%), Positives = 183/210 (86%)
Frame = +1
Query: 286 RYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMG 345
RY IYVKAGVYDEYIT+PK+AVNILMYGDGP KTIVTGRKN AG T TATF+NTA+G
Sbjct: 4 RYTIYVKAGVYDEYITIPKDAVNILMYGDGPGKTIVTGRKNGAAGVKTMQTATFANTALG 183
Query: 346 FIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGT 405
FIGKAMTFENTAGPAG QAVAFRN GDMSALVGCHI+GYQDTLYVQTNRQFYRNCVISGT
Sbjct: 184 FIGKAMTFENTAGPAGHQAVAFRNQGDMSALVGCHILGYQDTLYVQTNRQFYRNCVISGT 363
Query: 406 IDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALV 465
+DFIFGTSATLIQ STIIVR + N++N I ADGS + +NTGIVIQ CNI+P+AAL
Sbjct: 364 VDFIFGTSATLIQDSTIIVRMPS--PNQFNTITADGSYVNKLNTGIVIQGCNIVPKAALF 537
Query: 466 PEKFTVRSYLGRPWQNESKAVIMESTIGDF 495
P++FT++SYLGRPW+ +K V+MESTIG F
Sbjct: 538 PQRFTIKSYLGRPWKVLTKTVVMESTIG*F 627
Score = 24.6 bits (52), Expect(2) = 7e-92
Identities = 8/11 (72%), Positives = 9/11 (81%)
Frame = +2
Query: 492 IGDFIHQDGWT 502
+GDFIH GWT
Sbjct: 617 LGDFIHPXGWT 649
>TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precursor (EC
3.1.1.11) (Pectin methylesterase 3) (PE 3)., partial
(93%)
Length = 2088
Score = 318 bits (816), Expect = 3e-87
Identities = 200/582 (34%), Positives = 310/582 (52%), Gaps = 11/582 (1%)
Frame = +2
Query: 4 KVIISVVSLILVVGVIIGVVVDI---RKNGED-------PKVQ-TQQRNLRIMCQNAQDQ 52
++ I ++S I++V VII V I + N E P + T +L+ +C++ Q
Sbjct: 140 RITIIIISSIILVAVIIAAVAGILIHKHNTESSSSPNSLPNTELTPATSLKAVCESTQYP 319
Query: 53 KLCHDTLSSVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDDC 112
C ++SS+ ++ DP+ ++K A D + K +S +E + K A+ C
Sbjct: 320 NSCFSSISSLPDSNTTDPEQLFKLSLKVAIDELSK---LSLTRFSEKATEPRVKKAIGVC 490
Query: 113 KDLMQFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKK 172
+++ +LD L+ S + + D + D+ WLSA ++ C++ + N +
Sbjct: 491 DNVLADSLDRLNDSMSTIVDGGKMLSPAKIRDVETWLSAALTDHDTCLDAVGEVNSTAAR 670
Query: 173 IKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVTVDDQGYP 232
+ + + + + +L IV+ + +L NF++ RRLL +P
Sbjct: 671 GVIPEIERIMRNSTEFASNSLAIVSKVIRLLS----NFEVSNHHRRLLGE--------FP 814
Query: 233 SWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVK 292
W+ ++ R+LLA + + P+AVVA DGSGQ+KTI AL K + R+V+YVK
Sbjct: 815 EWLGTAERRLLATVVNE-----TVPDAVVAKDGSGQYKTIGEALKLVKKKSLQRFVVYVK 979
Query: 293 AGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMT 352
GVY E I + K N+++YGDG +T+V+G +N + GT T TATF+ GFI K +
Sbjct: 980 KGVYVENIDLDKNTWNVMIYGDGMTETVVSGSRNYIDGTPTFETATFAVKGKGFIAKDIQ 1159
Query: 353 FENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGT 412
F NTAG + QAVA R+ D S C VGYQDTLY +NRQFYR+C I+GTIDFIFG
Sbjct: 1160FLNTAGASKHQAVAMRSGSDQSVFYRCSFVGYQDTLYAHSNRQFYRDCDITGTIDFIFGN 1339
Query: 413 SATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVR 472
+A + Q+ I+ R+ N++N I A G N N+GIVIQ L +
Sbjct: 1340AAAVFQNCKIMPRQPM--SNQFNTITAQGKKDPNQNSGIVIQKSTF---TTLPGDNLIAP 1504
Query: 473 SYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGAN 532
+YLGRPW++ S +IM+S IG F+ GW +W P ++ +AEY NTGPGA+
Sbjct: 1505TYLGRPWKDFSTTIIMKSEIGSFLKPVGWISWVANVEPP-----SSILYAEYQNTGPGAD 1669
Query: 533 VARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEW 574
V RVKW GYK + +A K+T ++ GP+ P ++V++
Sbjct: 1670VPGRVKWAGYKPALGDEDAIKFTVDSFIQ-GPEWLPSASVQF 1792
>TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Citrus
sinensis}, partial (85%)
Length = 1993
Score = 312 bits (800), Expect = 2e-85
Identities = 208/603 (34%), Positives = 305/603 (50%), Gaps = 19/603 (3%)
Frame = +1
Query: 2 KGKVIISVVSLILVVGVIIGVVVDIRKNGEDPKVQTQQRN-----------LRIMCQNAQ 50
K K++IS+ + +L+V I+ +V KN K + + L+ C
Sbjct: 97 KKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTTL 276
Query: 51 DQKLCHDTLSSVRGAD--AADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMA 108
+LC +SS + K I+ ++ T V + E+L K+A
Sbjct: 277 YPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLTKREKIA 456
Query: 109 LDDCKDLMQFALDSL-DLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGF--DD 165
L DC + + LD L + N+ V + + ++ D++ +S+ I+ + C++GF DD
Sbjct: 457 LHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSHDD 636
Query: 166 ANDGEKKIKE--QFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDI-KPPSRRLLNS 222
A+ +K+ + Q HV+ + S A+A+ DI + N + +R+LL
Sbjct: 637 ADKEVRKVLQEGQIHVEHMCS----NALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEE 804
Query: 223 EVTVDDQGYPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKG 282
E V G+P WIS+ R+LL + ++ + VVA DGSG FKT+ A+A+ P
Sbjct: 805 ENGV---GWPEWISAGDRRLLQG-------STVKADVVVAADGSGNFKTVSEAVAAAPLK 954
Query: 283 NKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNT 342
+ RYVI +KAGVY E + VPK+ NI+ GDG TI+TG +N + G+ T ++AT +
Sbjct: 955 SSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIV 1134
Query: 343 AMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVI 402
F+ + +TF+NTAGPA QAVA R D+SA C I+ YQDTLYV NRQF+ NC I
Sbjct: 1135GGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFI 1314
Query: 403 SGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEA 462
SGT+DFIFG SA + Q+ I R+ N + N++ A G N NTGIVIQ C I
Sbjct: 1315SGTVDFIFGNSAVVFQNCDIHARRPN--SGQKNMVTAQGRVDPNQNTGIVIQKCRIGATK 1488
Query: 463 ALVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFA 522
L K +YLGRPW+ S+ V M+S+I D I GW W NT +
Sbjct: 1489DLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEW------NGNFALNTLVYR 1650
Query: 523 EYANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDLHVPH 582
EY NTGPGA ++RV WKG+K + S +EA T ++ WL P
Sbjct: 1651EYQNTGPGAGTSKRVTWKGFKVITSAAEAQSSTPGNFIGGS---------SWLGSTGFPF 1803
Query: 583 YLG 585
LG
Sbjct: 1804SLG 1812
>NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1
Length = 1698
Score = 295 bits (756), Expect = 3e-80
Identities = 206/587 (35%), Positives = 308/587 (52%), Gaps = 11/587 (1%)
Frame = +1
Query: 10 VSLILVVGVIIGVVVDIRKNGEDPK---VQTQQRNLRIMCQNAQDQKLCHDTLSSVRGAD 66
VS IL+V ++ V V + K G+ + + Q+N+ ++CQ+ + ++ CH TL A
Sbjct: 61 VSCILLVAMVGVVAVSLTKGGDGEQKAHISNSQKNVDMLCQSTKFKETCHKTLEK---AS 231
Query: 67 AADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDDCKDLMQFALDSLDLS 126
++ K I A+ A + + K N S L E + K A++ C +++ +A+D + S
Sbjct: 232 FSNMKNRIKGALGATEEELRKHINNSA-LYQELATDSMTKQAMEICNEVLDYAVDGIHKS 408
Query: 127 NNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDAND--GEKKIKEQFHVQSLYS 184
+ + + + D++ WL+ +S++Q C++GF + GE K L +
Sbjct: 409 VGTLDQFDFHKLSEYAFDIKVWLTGTLSHQQTCLDGFVNTKTHAGETMAKV------LKT 570
Query: 185 VQKVTAVALDIVTGLSDILQQFNLN-FDIKPPSRRLLNSEVTVDDQGYPSWISSSGRKLL 243
++++ A+D++ +S IL+ F+ + + + SRRLL+ D G PSW+S R LL
Sbjct: 571 SMELSSNAIDMMDVVSRILKGFHPSQYGV---SRRLLS------DDGIPSWVSDGHRHLL 723
Query: 244 AKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEYITVP 303
A N++ NAVVA DGSGQFKT+ AL + P N +VIYVKAGVY E + V
Sbjct: 724 AG-------GNVKANAVVAQDGSGQFKTLTDALKTVPPTNAAPFVIYVKAGVYKETVNVA 882
Query: 304 KEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENTAGPAGMQ 363
KE + + GDGP KT TG N G NT TATF F+ K + FENTAG + Q
Sbjct: 883 KEMNYVTVIGDGPTKTKFTGSLNYADGINTYKTATFGVNGANFMAKDIGFENTAGTSKFQ 1062
Query: 364 AVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLIQSSTII 423
AVA R D + C + G+QDTL+V++ RQFYR+C ISGTIDF+FG + + Q+ +I
Sbjct: 1063AVALRVTADQAIFHNCQMDGFQDTLFVESQRQFYRDCAISGTIDFVFGDAFGVFQNCKLI 1242
Query: 424 VR---KGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAAL--VPEKFTVRSYLGRP 478
R KG + ++ A G N + +V + E AL V K SYLGRP
Sbjct: 1243CRVPAKG-----QKCLVTAGGRDKQNSASALVFLSSHFTGEPALTSVTPKL---SYLGRP 1398
Query: 479 WQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARRVK 538
W+ SK VIM+STI +G+ K +TC F EY N GPGA+ RVK
Sbjct: 1399WKLYSKVVIMDSTIDAMFAPEGYMPMVGGAFK------DTCTFYEYNNKGPGADTNLRVK 1560
Query: 539 WKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDLHVPHYLG 585
W G K V++ + A +Y + + TA + W+ VP+ LG
Sbjct: 1561WHGVK-VLTSNVAAEYYPGKFFEIVNATARDT---WIVKSGVPYSLG 1689
>TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11)
precursor - garden pea, complete
Length = 1860
Score = 295 bits (755), Expect = 3e-80
Identities = 177/529 (33%), Positives = 281/529 (52%), Gaps = 13/529 (2%)
Frame = +2
Query: 45 MCQNAQDQKLCHDTLSSV-RGADAADPKAY----IAAAVKAATDNVIKAFNMSERLTTEY 99
+C++A D K C +S V +G+ ++ K + + + + +T ++ KA + + +
Sbjct: 194 LCEHALDTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRV 373
Query: 100 GKKNGAKMALDDCKDLMQFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGC 159
++AL+DC++LM ++D + S + NNI++ HD WLS+V++ C
Sbjct: 374 NSPR-EEIALNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDA----HTWLSSVLTNHATC 538
Query: 160 MEGFDDANDGEKKIKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRR- 218
++G + G ++ + + L S + + L + PP
Sbjct: 539 LDGLE----GSSRVVMESDLHDLISRARSSLAVL----------------VSVLPPKAND 658
Query: 219 -LLNSEVTVDDQGYPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALA 277
++ ++ D +PSW++S R+LL +I+ N VVA DGSG+FKT+ A+A
Sbjct: 659 GFIDEKLNGD---FPSWVTSKDRRLLESSV-----GDIKANVVVAQDGSGKFKTVAQAVA 814
Query: 278 SYPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTA 337
S P K RYVIYVK G Y E I + K+ N+++ GDG TI+TG N + GT T +A
Sbjct: 815 SAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSA 994
Query: 338 TFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFY 397
T + GFI + + F+NTAGP QAVA R D S + C I +QDTLY +NRQFY
Sbjct: 995 TVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFY 1174
Query: 398 RNCVISGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCN 457
R+ I+GT+DFIFG +A + Q S + RK N+ N++ A G N NT IQ C+
Sbjct: 1175RDSYITGTVDFIFGNAAVVFQKSKLAARKPM--ANQKNMVTAQGREDPNQNTATSIQQCD 1348
Query: 458 IIPEAALVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHEN 517
+IP + L P + ++++YLGRPW+ S+ V+++S + I GW W + K
Sbjct: 1349VIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEW----DAASKDFLQ 1516
Query: 518 TCYFAEYANTGPGANVARRVKWKGYKGVISRSEATKYT------ASIWL 560
T Y+ EY N+G GA +RV W GY + + +EA+K+T ++WL
Sbjct: 1517TLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWL 1663
>TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, complete
Length = 1981
Score = 279 bits (713), Expect = 2e-75
Identities = 175/528 (33%), Positives = 271/528 (51%), Gaps = 12/528 (2%)
Frame = +3
Query: 45 MCQNAQDQKLCHDTLSSV-RGADAADPKAY----IAAAVKAATDNVIKAFNMSERLTTEY 99
+C++A D C ++ V +G+ + K + + + + +T ++ KA + + +
Sbjct: 201 VCEHAVDTNSCLTHVAEVVQGSTLDNTKDHKLSTLISLLTKSTTHIRKAMDTANVIKRRI 380
Query: 100 GKKNGAKMALDDCKDLMQFALDSL-DLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQG 158
+ AL+ C+ LM +++ + D +DN D D WLS+V++
Sbjct: 381 NSPR-EENALNVCEKLMNLSMERVWDSVLTLTKDNM-----DSQQDAHTWLSSVLTNHAT 542
Query: 159 CMEGFDDANDGEKKIKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRR 218
C++G + G + + +Q L + + + L V D
Sbjct: 543 CLDGLE----GTSRAVMENDIQDLIARARSSLAVLVAVLPPKD--------------HDE 668
Query: 219 LLNSEVTVDDQGYPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALAS 278
++ + D +PSW++S R+LL +++ N VVA DGSG+FKT+ A+AS
Sbjct: 669 FIDESLNGD---FPSWVTSKDRRLLESSV-----GDVKANVVVAKDGSGKFKTVAEAVAS 824
Query: 279 YPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTAT 338
P RYVIYVK G+Y E + + N+++ GDG TI+TG N + GT T TAT
Sbjct: 825 APNKGTARYVIYVKKGIYKENVEIASSKTNVMLLGDGMDATIITGSLNYVDGTGTFQTAT 1004
Query: 339 FSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYR 398
+ FI + + F+NTAGP QAVA R D S + C I +QDTLY TNRQFYR
Sbjct: 1005VAAVGDWFIAQDIGFQNTAGPQKHQAVALRVGSDRSVINRCKIDAFQDTLYAHTNRQFYR 1184
Query: 399 NCVISGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNI 458
+ I+GTIDFIFG +A ++Q ++ RK + N N++ A G N NT IQ C++
Sbjct: 1185DSFITGTIDFIFGDAAVVLQKCKLVARKPMANQN--NMVTAQGRIDPNQNTATSIQQCDV 1358
Query: 459 IPEAALVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENT 518
IP L P +V++YLGRPW+ S+ V+M+S +G I GW W + K T
Sbjct: 1359IPSTDLKPVIGSVKTYLGRPWKKYSRTVVMQSLLGAHIDPTGWAEW----DAASKDFLQT 1526
Query: 519 CYFAEYANTGPGANVARRVKWKGYKGVISRSEATKYT------ASIWL 560
Y+ EY N+GPGA ++RVKW GY +I+ +EA K+T ++WL
Sbjct: 1527LYYGEYMNSGPGAGTSKRVKWPGYH-IINTAEANKFTVAQLIQGNVWL 1667
>TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, partial (98%)
Length = 2029
Score = 277 bits (709), Expect = 7e-75
Identities = 171/528 (32%), Positives = 267/528 (50%), Gaps = 11/528 (2%)
Frame = +2
Query: 44 IMCQNAQDQKLCHDTLSSVRGADAA----DPKAYIAAAVKA-ATDNVIKAFNMSERLTTE 98
+ C++A D K C +S V D ++ ++ +T ++ A + + +
Sbjct: 206 VYCEHAVDTKSCLAHVSEVSHVPTLVTTKDQNLHVLLSLLTKSTTHIQNAMDTASVIKRR 385
Query: 99 YGKKNGAKMALDDCKDLMQFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQG 158
++AL DC+ LM +++ + + + NNI D D WLS+V++
Sbjct: 386 INSPR-EEIALSDCEQLMDLSMNRIWDTMLKLTKNNI----DSQQDAHTWLSSVLTNHAT 550
Query: 159 CMEGFDDANDGEKKIKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRR 218
C++G + G ++ + +Q L S + + +V D R
Sbjct: 551 CLDGLE----GSSRVVMENDLQDLISRARSSLAVFLVVFPQKD---------------RD 673
Query: 219 LLNSEVTVDDQGYPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALAS 278
E + + +PSW++S R+LL +I+ N VVA DGSG+FKT+ A+AS
Sbjct: 674 QFIDETLIGE--FPSWVTSKDRRLLETAV-----GDIKANVVVAQDGSGKFKTVAEAVAS 832
Query: 279 YPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTAT 338
P K +YVIYVK G Y E + + + N+++ GDG TI+TG N + GT T ++T
Sbjct: 833 APDNGKTKYVIYVKKGTYKENVEIGSKKTNVMLVGDGMDATIITGNLNFIDGTTTFKSST 1012
Query: 339 FSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYR 398
+ GFI + + F+N AG A QAVA R D S + C I +QDTLY +NRQFYR
Sbjct: 1013VAAVGDGFIAQDIWFQNMAGAAKHQAVALRVGSDQSVINRCRIDAFQDTLYAHSNRQFYR 1192
Query: 399 NCVISGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNI 458
+ VI+GTIDFIFG +A + Q ++ RK + N N+ A G NTG IQ C++
Sbjct: 1193DSVITGTIDFIFGNAAVVFQKCKLVARKPMANQN--NMFTAQGREDPGQNTGTSIQQCDL 1366
Query: 459 IPEAALVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENT 518
P + L P +++++LGRPW+ S+ V+M+S + I GW W + K T
Sbjct: 1367TPSSDLKPVVGSIKTFLGRPWKKYSRTVVMQSFLDSHIDPTGWAEW----DAASKDFLQT 1534
Query: 519 CYFAEYANTGPGANVARRVKWKGYKGVISRSEATKYT------ASIWL 560
Y+ EY N GPGA A+RV W GY + + +EA+K+T ++WL
Sbjct: 1535LYYGEYLNNGPGAGTAKRVTWPGYHVINTAAEASKFTVAQLIQGNVWL 1678
>TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 {Arabidopsis
thaliana}, partial (79%)
Length = 2442
Score = 259 bits (663), Expect = 2e-69
Identities = 165/455 (36%), Positives = 234/455 (51%), Gaps = 5/455 (1%)
Frame = +3
Query: 106 KMALDDCKDLMQFALDSLDLSN--NCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGF 163
K A DC +L F + L + C + + + WLS ++ + C GF
Sbjct: 843 KAAWADCLELYDFTVQKLSQTKYTKCTQ-----------YESQTWLSTALTNLETCKNGF 989
Query: 164 DDANDGEKKIKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKP---PSRRLL 220
D VT L +++ L L+ + P PS +
Sbjct: 990 YDLG--------------------VTNYVLPLLSNNVTKLLSNTLSLNKVPYQQPSYK-- 1103
Query: 221 NSEVTVDDQGYPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYP 280
G+P+W+ RKLL A + N VVA DGSG++ T++AA + P
Sbjct: 1104 --------DGFPTWVKPGDRKLLQTSS-----AASKANVVVAKDGSGKYTTVKAATDAAP 1244
Query: 281 KGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFS 340
G+ RYVIYVKAGVY+E + + +A N+++ GDG KTI+TG K+ GT T +AT +
Sbjct: 1245 SGS-GRYVIYVKAGVYNEQVEI--KAKNVMLVGDGIGKTIITGSKSVGGGTTTFRSATVA 1415
Query: 341 NTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNC 400
T GFI + +TF NTAG A QAVAFR+ D+S C GYQDTLYV + RQFYR C
Sbjct: 1416 ATGDGFIAQDITFRNTAGAANHQAVAFRSGSDLSVFYKCSFEGYQDTLYVHSERQFYREC 1595
Query: 401 VISGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIP 460
I GT+DFIFG +A ++Q+ I R + + A G N NTGI+I + +
Sbjct: 1596 NIYGTVDFIFGNAAVVLQNCNIFARN---PPAKTITVTAQGRTDPNQNTGIIIHNSRVSA 1766
Query: 461 EAALVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCY 520
++ L P +V+SYLGRPWQ S+ V M++ + FI+ GW W +T Y
Sbjct: 1767 QSDLNPS--SVKSYLGRPWQKYSRTVFMKTVLDGFINPAGWLPW------DGNFALDTLY 1922
Query: 521 FAEYANTGPGANVARRVKWKGYKGVISRSEATKYT 555
+AEYANTG G++ + RV WKGY + S S+A+ +T
Sbjct: 1923 YAEYANTGSGSSTSNRVTWKGYHVLTSASQASPFT 2027
>TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylesterase isoform
alpha {Vigna radiata}, partial (94%)
Length = 1655
Score = 246 bits (629), Expect = 1e-65
Identities = 132/322 (40%), Positives = 186/322 (56%)
Frame = +2
Query: 234 WISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKA 293
W+S RKLL + + + VVA DG+G F TI A+A+ P + R+VI++KA
Sbjct: 41 WVSPKDRKLLQAAVNQ-----TKFDLVVAKDGTGNFATIGEAIAAAPNSSATRFVIHIKA 205
Query: 294 GVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTF 353
G Y E + V K+ N+++ GDG +T+V +N + G T ++TF+ FI K +TF
Sbjct: 206 GAYFENVEVIKKKTNLMLVGDGIGQTVVKASRNVVDGWTTFQSSTFAVVGDKFIAKGITF 385
Query: 354 ENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTS 413
EN+AGP+ QAVA RN D SA C V YQDTLYV + RQFYR C + GT+DFIFG +
Sbjct: 386 ENSAGPSKHQAVAVRNGADFSAFYQCSFVAYQDTLYVHSLRQFYRECDVYGTVDFIFGNA 565
Query: 414 ATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRS 473
A + Q+ + RK D + N+ A G N NTGI I +C I A L+P + T +S
Sbjct: 566 AVVFQNCNLYARKP--DPKQKNLFTAQGREDPNQNTGISILNCKIAAAADLIPVQSTFKS 739
Query: 474 YLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANV 533
YLGRPW+ S+ V + S I + I GW W +T ++ EY N GPG+N
Sbjct: 740 YLGRPWKKYSRTVYLNSFIDNLIDPPGWLEW------NGTFALDTLFYGEYKNRGPGSNT 901
Query: 534 ARRVKWKGYKGVISRSEATKYT 555
+ RV W GYK + + +EA+++T
Sbjct: 902 SARVTWPGYKVITNATEASQFT 967
>TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1929
Score = 238 bits (608), Expect = 4e-63
Identities = 160/491 (32%), Positives = 237/491 (47%), Gaps = 17/491 (3%)
Frame = +3
Query: 105 AKMALDDCKDLMQFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFD 164
A AL DC+ L D L S V Q +++ LSA+++ +Q C++G
Sbjct: 306 ALRALQDCQSLSDLNFDFLSSSFQTVNKTTKFLPSLQGENIQTLLSAILTNQQTCLDGLK 485
Query: 165 DANDGEKKIKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEV 224
D + S +S + + L T L + F + P + +
Sbjct: 486 DTS-------------SAWSFRNGLTIPLSNDTKLYSVSLAFFTKGWVNPKTNKTSFPNS 626
Query: 225 TVDDQGYPS--------------WISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFK 270
++G+ + + S S RKLL Q G +R V+ DGSG F
Sbjct: 627 KHSNKGFKNGRLPLKMTSKTRAIYESVSRRKLLQSNQ-VGEDVVVRDIVTVSQDGSGNFT 803
Query: 271 TIQAALASYPK---GNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQ 327
TI A+A+ P + ++IYV AGVY+EYIT+ K+ ++M GDG KTI+TG +
Sbjct: 804 TINDAIAAAPNKSVSSDGYFLIYVTAGVYEEYITIDKKKTYLMMIGDGINKTIITGNHSV 983
Query: 328 MAGTNTQNTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDT 387
+ G T + TF+ GF+G MT NTAG QAVA RN D+S C GYQDT
Sbjct: 984 VDGWTTFGSPTFAVVGQGFVGVNMTIRNTAGAVKHQAVALRNGADLSTFYSCSFEGYQDT 1163
Query: 388 LYVQTNRQFYRNCVISGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNM 447
LY + RQFYR C I GT+DFIFG + + Q+ + R ++N I A G N
Sbjct: 1164LYTHSLRQFYRECDIYGTVDFIFGNAKVVFQNCNLYPRLPM--SGQFNAITAQGRTDPNQ 1337
Query: 448 NTGIVIQDCNIIPEAALVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEE 507
+TG I +C I L V +YLGRPW+ S+ V M++ + + I+ GW W E
Sbjct: 1338DTGTSIHNCTIKATDDLAASNGAVSTYLGRPWKEYSRTVYMQTFMDNVINVAGWRAWDGE 1517
Query: 508 QNKPEKHHENTCYFAEYANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTA 567
+T Y+AE+ N+GPG++ RV W+GY VI+ ++A +T + +L G
Sbjct: 1518------FALSTLYYAEFNNSGPGSSTDGRVTWQGYH-VINATDAANFTVANFL-LGDDWL 1673
Query: 568 PKSAVEWLHDL 578
P++ V + + L
Sbjct: 1674PQTGVSYTNSL 1706
>TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectinesterase
{Arabidopsis thaliana}, partial (59%)
Length = 1963
Score = 230 bits (587), Expect = 1e-60
Identities = 168/520 (32%), Positives = 259/520 (49%), Gaps = 23/520 (4%)
Frame = +2
Query: 46 CQNAQDQKLCHDTLSSVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGA 105
C+ KLC LS++R + + DP Y ++K +K E++ ++ ++ +
Sbjct: 155 CKTTLYPKLCRSMLSAIRSSPS-DPYNYGKFSIKQN----LKVARKLEKVFIDFLNRHQS 319
Query: 106 KMALD--------DCKDLMQFALDSLDLSNNCVRDNNIEAVHDQTA---DMRNWLSAVIS 154
+L+ DCKDL +D L+ ++ ++ + + T + ++LSAV +
Sbjct: 320 SSSLNHEEVGALVDCKDLNSLNVDYLESISDELKSASSSSSSSDTELVDKIESYLSAVAT 499
Query: 155 YKQGCMEGF-----DDANDGEKKIKE--QFHVQSLYSVQKVTAVALDIVTGLSDILQQFN 207
C +G + AN +K+ QF+ SL V + A++ ++ + N
Sbjct: 500 NHYTCYDGLVVTKSNIANALAVPLKDATQFYSVSLGLVTE--ALSKNMKRNKTRKHGLPN 673
Query: 208 LNFDIKPPSRRLLNSEVTVDDQGYPSWISSSGR--KLLAKMQRKGWRANIRPNAVVANDG 265
+F ++ P +L+ T S +S R ++L + + G N +V+ G
Sbjct: 674 KSFKVRQPLEKLIKLLRTKYSCQKTSSNCTSTRTERILKESESHGILLN--DFVLVSPYG 847
Query: 266 SGQFKTIQAALASYPKGNKDR---YVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVT 322
+I A+A+ P K Y+IYV+ G Y+EY+ VPK NIL+ GDG TI+T
Sbjct: 848 IANHTSIGDAIAAAPNNTKPEDGYYLIYVREGYYEEYVIVPKHKNNILLVGDGINNTIIT 1027
Query: 323 GRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIV 382
G + + G T N++TF+ + FI +TF NTAGP QAVA RN D+S C
Sbjct: 1028GNHSVIDGWTTFNSSTFAVSGERFIAVDITFRNTAGPEKHQAVAVRNNADLSTFYRCSFE 1207
Query: 383 GYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGS 442
GYQDTLYV + RQFYR+C I GT+DFIFG +A + Q+ I RK N+ N + A G
Sbjct: 1208GYQDTLYVHSVRQFYRDCKIYGTVDFIFGNAAVVFQNCNIYARKPL--PNQKNAVTAQGR 1381
Query: 443 PLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWT 502
N NTGI IQ+C I L + + SYLGRPW+ S+ V M+S IGDF+ GW
Sbjct: 1382TDPNQNTGISIQNCTIDAAQDLANDLNSTMSYLGRPWKIYSRTVYMQSYIGDFVQPSGWL 1561
Query: 503 TWPEEQNKPEKHHENTCYFAEYANTGPGANVARRVKWKGY 542
W +T ++ E+ N GPG+ RV+W G+
Sbjct: 1562EWNGTVGL------DTIFYGEFNNYGPGSVTNNRVQWPGH 1663
>TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectinesterase
{Arabidopsis thaliana}, partial (34%)
Length = 691
Score = 170 bits (430), Expect = 2e-42
Identities = 98/218 (44%), Positives = 127/218 (57%)
Frame = +3
Query: 232 PSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYV 291
P+W+ RKLL K +A+I VVA DGSG++KTI AAL P + R VIYV
Sbjct: 54 PNWLHHKDRKLLQKDSDLKKKADI----VVAKDGSGKYKTISAALKHVPNKSDKRTVIYV 221
Query: 292 KAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAM 351
K G+Y E + V K N+++ GDG TIV+G N + GT T +TATF+ FI + +
Sbjct: 222 KKGIYYENVRVEKTKWNVMIIGDGMNVTIVSGSLNFIDGTPTFSTATFAVFGRNFIARDI 401
Query: 352 TFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFG 411
F+NTAGP QAVA D + C + YQDTLY +NRQFYR C I GT+DFIFG
Sbjct: 402 GFKNTAGPQKHQAVALMTSADQAVFYKCSMDAYQDTLYAHSNRQFYRECNIYGTVDFIFG 581
Query: 412 TSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNT 449
SA ++Q+ I+ R+ + N I A G NMNT
Sbjct: 582 NSAVVLQNCNILPRQPM--PGQQNTITAQGKTDPNMNT 689
>BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like protein -
Arabidopsis thaliana, partial (36%)
Length = 658
Score = 154 bits (388), Expect = 1e-37
Identities = 92/217 (42%), Positives = 122/217 (55%), Gaps = 9/217 (4%)
Frame = +1
Query: 273 QAALASYPKGNKDR--YVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQ-MA 329
Q A+ + P DR +VIY+K GVY+E + VP + N++ GDG KT++TG N
Sbjct: 1 QEAVNAAPDNGVDRKRFVIYIKEGVYEETVRVPLKKRNVVFLGDGIGKTVITGSANVGQP 180
Query: 330 GTNTQNTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLY 389
G T N+AT + GF+ K +T ENTAGP QAVAFR D+S + C +G QDTLY
Sbjct: 181 GMTTYNSATVAVLGDGFMAKDLTIENTAGPDAHQAVAFRLDSDLSVIENCEFLGNQDTLY 360
Query: 390 VQTNRQFYRNCVISGTIDFIFGTSATLIQSSTIIVRKGNY--DHNEYNVIVADGSPLVNM 447
+ RQFY++C I G +DFIFG SA + Q I+VR + E N I A G
Sbjct: 361 AHSLRQFYKSCRIVGNVDFIFGNSAAIFQDCQILVRPRQLKPEKGENNAITAHGRTDPAQ 540
Query: 448 NTGIVIQDCNI---IPEAALVPEKFTV-RSYLGRPWQ 480
+TG V Q+C I AL V ++YLGRPW+
Sbjct: 541 STGFVFQNCLINGTEDYMALYHSNPKVHKNYLGRPWK 651
>TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor (EC
3.1.1.11) (Pectin methylesterase) (PE) (P65). [Alfalfa],
partial (53%)
Length = 732
Score = 147 bits (370), Expect(2) = 6e-37
Identities = 88/216 (40%), Positives = 119/216 (54%), Gaps = 3/216 (1%)
Frame = +2
Query: 342 TAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCV 401
T+ F + FEN+AG A QAVA R D + C + GYQDTLY Q+ RQFYR+C
Sbjct: 74 TSAHFTALNVGFENSAGAAKHQAVALRVTADKALFYNCQMNGYQDTLYTQSKRQFYRDCT 253
Query: 402 ISGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPE 461
I+GTIDF+FG + + Q+ +IVRK N+ ++ A G V+ + +V Q+C+ E
Sbjct: 254 ITGTIDFVFGDAVGVFQNCKLIVRKPM--ANQQCMVTAGGRTKVDSVSALVFQNCHFTGE 427
Query: 462 AALVPEKFTVR---SYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENT 518
PE T++ +YLGRPW+N SK VI++S I +G+ W K T
Sbjct: 428 ----PEVLTMQPKIAYLGRPWRNFSKVVIVDSLIDGLFVPEGYMPWMGNLFK------ET 577
Query: 519 CYFAEYANTGPGANVARRVKWKGYKGVISRSEATKY 554
C + EY N G GA +VKW G K IS EA KY
Sbjct: 578 CTYLEYNNKGAGAATNLKVKWPGVK-TISAGEAAKY 682
Score = 25.8 bits (55), Expect(2) = 6e-37
Identities = 12/21 (57%), Positives = 13/21 (61%)
Frame = +1
Query: 320 IVTGRKNQMAGTNTQNTATFS 340
I TG K+ G T NTATFS
Sbjct: 7 IFTGSKSYADGVQTYNTATFS 69
>BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precursor (EC
3.1.1.11) (Pectin methylesterase) (PE). [Peach] {Prunus
persica}, partial (36%)
Length = 589
Score = 149 bits (375), Expect = 4e-36
Identities = 71/162 (43%), Positives = 106/162 (64%)
Frame = +2
Query: 265 GSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGR 324
GSG + + A+++ P+ + RYVIYVK GVY E + + K+ NI++ G+G TI++G
Sbjct: 2 GSGDYAKVMDAVSAAPESSMKRYVIYVKKGVYVENVEIKKKKWNIMLIGEGMDATIISGS 181
Query: 325 KNQMAGTNTQNTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGY 384
+N + G+ T +ATF+ + GFI + ++F+NTAG QAVA R+ D+S C I GY
Sbjct: 182 RNYVDGSTTFRSATFAVSGRGFIARDISFQNTAGAEKHQAVALRSDSDLSVFYRCGIFGY 361
Query: 385 QDTLYVQTNRQFYRNCVISGTIDFIFGTSATLIQSSTIIVRK 426
QD+LY T RQFYR C ISGT+DFIFG + + Q+ I+ ++
Sbjct: 362 QDSLYTHTMRQFYRECKISGTVDFIFGDATAVFQNCQILAKE 487
>AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (25%)
Length = 495
Score = 127 bits (319), Expect = 1e-29
Identities = 67/144 (46%), Positives = 89/144 (61%), Gaps = 3/144 (2%)
Frame = +1
Query: 264 DGSGQFKTIQAALASYPKGN--KDRY-VIYVKAGVYDEYITVPKEAVNILMYGDGPAKTI 320
DGSG F TI A+A+ P D Y +I++ GVY+EY+++ K+ ++M G+G +TI
Sbjct: 64 DGSGNFTTINDAIAAAPNNTVASDGYFLIFITEGVYEEYVSIDKKKKYLMMIGEGINQTI 243
Query: 321 VTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCH 380
+TG +N G T N+ATF+ A GF+ +TF NTAG A QAVA R+ DMS C
Sbjct: 244 ITGNRNVADGFTTFNSATFAVVAQGFVAVNITFRNTAGAAKNQAVALRSGADMSTFYSCS 423
Query: 381 IVGYQDTLYVQTNRQFYRNCVISG 404
GYQDTLY + RQFYR C I G
Sbjct: 424 FEGYQDTLYTHSLRQFYRECDIYG 495
>TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pectin
methylesterase {Arabidopsis thaliana}, partial (33%)
Length = 1289
Score = 102 bits (253), Expect(2) = 2e-26
Identities = 77/229 (33%), Positives = 102/229 (43%), Gaps = 10/229 (4%)
Frame = +3
Query: 261 VANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTI 320
V+ DGS QFK+I AL S N R +I + G Y E I VPK I GD
Sbjct: 243 VSQDGSAQFKSITEALNSIQPYNIRRVIISIAPGYYREKIVVPKTLPFITFLGDVRDPPT 422
Query: 321 VTGRKNQM------AGTNTQNTATFSNTAMGFIGKAMTFENTAG-PAGM---QAVAFRNI 370
+TG Q A T N+AT + A F+ + FENTA P G QAVA R
Sbjct: 423 ITGNDTQSVTGSDGAQLRTFNSATVAVNASYFMAININFENTASFPIGSKVEQAVAVRIT 602
Query: 371 GDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLIQSSTIIVRKGNYD 430
G+ +A C G QDTLY ++ NC I G++DFI G +L + TI
Sbjct: 603 GNKTAFYNCTFSGVQDTLYDHKGLHYFNNCTIKGSVDFICGHGKSLYEGCTI-----RSI 767
Query: 431 HNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLGRPW 479
N I A + ++G ++ +I + +YLGRPW
Sbjct: 768 ANNMTSITAQSGSNPSYDSGFSFKNSMVIGDGP---------TYLGRPW 887
Score = 35.8 bits (81), Expect(2) = 2e-26
Identities = 22/81 (27%), Positives = 39/81 (47%)
Frame = +1
Query: 481 NESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARRVKWK 540
N S+ V + + + + GW W + K + N Y+ EY +GPG+N A RV W
Sbjct: 892 NYSQVVFSYTYMDNSVLPKGWEDWNDT-----KRYMNA-YYGEYKCSGPGSNTAGRVPW- 1050
Query: 541 GYKGVISRSEATKYTASIWLD 561
+++ EA + + ++D
Sbjct: 1051--ARMLNDKEAQVFIGTQYID 1107
>TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE8B
precursor (EC 3.1.1.11) (Pectin methylesterase) (PE).
[Peach] {Prunus persica}, partial (34%)
Length = 1415
Score = 110 bits (276), Expect = 1e-24
Identities = 85/297 (28%), Positives = 141/297 (46%), Gaps = 16/297 (5%)
Frame = +1
Query: 59 LSSVRGADAADPKAYIAAAVKAAT-------DNVI----KAFNMSERLTTEYGKKNGAKM 107
LS A+ P ++ ++++ +T + VI K ++ R T +G +
Sbjct: 148 LSLPHSTFASSPNDFVGSSLRVSTAKFANSAEEVITVLQKVISILSRFTNVFGHSRTSN- 324
Query: 108 ALDDCKDLMQFALDSLDLSNNCV-----RDNNIEAVHDQTADMRNWLSAVISYKQGCMEG 162
A+ DC DL+ +LD L+ S + +DN+ ++ D+R WLSAV+ Y C+EG
Sbjct: 325 AVSDCLDLLDMSLDQLNQSISAAQKPKEKDNSTGKLN---CDLRTWLSAVLVYPDTCIEG 495
Query: 163 FDDANDGEKKIKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNS 222
+ + HV SL V + ++V+G D
Sbjct: 496 LEGSIVKGLISSGLDHVMSL-----VANLLGEVVSGNDD--------------------- 597
Query: 223 EVTVDDQGYPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKG 282
++ + +PSWI KLL Q G + +AVVA DGSG + + A+++ P+
Sbjct: 598 QLATNKDRFPSWIRDEDTKLL---QANG----VTADAVVAADGSGDYAKVMDAVSAAPES 756
Query: 283 NKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATF 339
+ RYVIYVK GVY E + + K+ NI++ G+G TI++G +N + G+ T +ATF
Sbjct: 757 SMKRYVIYVKKGVYVENVEIKKKKWNIMLIGEGMDATIISGSRNYVDGSTTFRSATF 927
>TC91054 similar to GP|20521423|dbj|BAB91933. putative pectin methylesterase
{Oryza sativa (japonica cultivar-group)}, partial (42%)
Length = 809
Score = 88.6 bits (218), Expect = 6e-18
Identities = 64/175 (36%), Positives = 89/175 (50%), Gaps = 16/175 (9%)
Frame = +1
Query: 260 VVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKT 319
V + G F +IQAA+ S P N R VI V AGVY E +++P I + G G KT
Sbjct: 289 VYKHSSKGGFSSIQAAIDSLPFINLVRVVIKVHAGVYTEKVSIPALKSFITIQGAGADKT 468
Query: 320 IV----------TGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENTA-----GPAGMQA 364
IV G K Q G T +ATF+ + FI K +TF+NTA G Q
Sbjct: 469 IVQWGDTALTPNPGAKGQTLG--TYGSATFAVNSPYFIAKNITFKNTAPIPKPEAVGKQG 642
Query: 365 VAFRNIGDMSALVGCHIV-GYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLIQ 418
VA R D + ++G I+ G T Y R +Y++ I G++DFIFG + +L++
Sbjct: 643 VALRISADTAMVLGLQILRGTGHTWYDHIGRHYYKDWYIEGSVDFIFGNALSLLK 807
>TC92167 similar to PIR|T05203|T05203 pectinesterase homolog F4I10.160 -
Arabidopsis thaliana, partial (11%)
Length = 652
Score = 76.3 bits (186), Expect = 3e-14
Identities = 56/152 (36%), Positives = 72/152 (46%), Gaps = 8/152 (5%)
Frame = +1
Query: 353 FENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFY----RNCVISGTIDF 408
F NTAGP G QAVA R D + C G+QDTLY +RQF+ N DF
Sbjct: 220 FRNTAGPEGHQAVAARVQADRAVFANCRFEGFQDTLYTVAHRQFFP*LHHN---REQFDF 390
Query: 409 IFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPE- 467
IFG + L + + K N+ + V + T + Q I L+
Sbjct: 391 IFGDPSPLYSKTASWLLK-----NQVSAKVTR*QHKEDWTTNKIQQLFYINVHLKLMTHW 555
Query: 468 ---KFTVRSYLGRPWQNESKAVIMESTIGDFI 496
K TV+SYLGR W+ S+ V+MES IGDFI
Sbjct: 556 FLLKATVKSYLGRAWKQFSRTVVMESDIGDFI 651
Score = 48.1 bits (113), Expect(2) = 3e-07
Identities = 47/183 (25%), Positives = 78/183 (41%), Gaps = 11/183 (6%)
Frame = +3
Query: 297 DEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENT 356
DE +T+ + +I +YGDG K+I+TG KN G NTA+ GF+G A
Sbjct: 51 DEVVTITDKMKDITIYGDGSQKSIITGSKNFRDGVTNINTASLVVLGEGFLGLA------ 212
Query: 357 AGPAGMQAVAF----RNIGDMSALVGCHIVGY------QDTLYVQTNRQFYRNCVISGTI 406
+G+Q ++ + S C + + ++ T GTI
Sbjct: 213 ---SGIQKHSWS*RTSSSSS*SPSRSCCVCQLPF*RLPRHAIHSGT*TILPVAAS*QGTI 383
Query: 407 DF-IFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALV 465
F ++ + Q+ ++V+K + + N + A G NT IV+ C + + ALV
Sbjct: 384 RFHLW*PQPVIFQNCILVVKKPSV--GQSNAVTAQGRLDNKQNTAIVLHKCTLKADDALV 557
Query: 466 PEK 468
P K
Sbjct: 558 PVK 566
Score = 24.6 bits (52), Expect(2) = 3e-07
Identities = 10/11 (90%), Positives = 10/11 (90%)
Frame = +2
Query: 286 RYVIYVKAGVY 296
RYVIYVK GVY
Sbjct: 17 RYVIYVKEGVY 49
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,547,064
Number of Sequences: 36976
Number of extensions: 206738
Number of successful extensions: 1058
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 996
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1000
length of query: 588
length of database: 9,014,727
effective HSP length: 101
effective length of query: 487
effective length of database: 5,280,151
effective search space: 2571433537
effective search space used: 2571433537
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144563.4


