

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144563.2 + phase: 0
(576 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
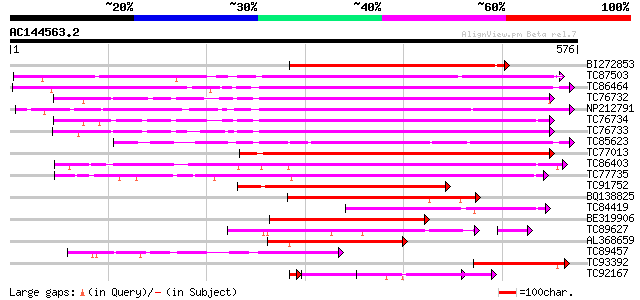
Score E
Sequences producing significant alignments: (bits) Value
BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-... 365 e-101
TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precurs... 343 8e-95
TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Cit... 335 3e-92
TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.... 322 2e-88
NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1 321 6e-88
TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 308 4e-84
TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 301 6e-82
TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 ... 300 8e-82
TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylestera... 278 5e-75
TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF028... 270 1e-72
TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectineste... 265 3e-71
TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectineste... 194 6e-50
BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like pr... 167 8e-42
TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor ... 157 8e-39
BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precurso... 153 2e-37
TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pec... 109 7e-31
AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886... 127 1e-29
TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE... 119 3e-27
TC93392 similar to PIR|T49241|T49241 pectinesterase-like protein... 102 4e-22
TC92167 similar to PIR|T05203|T05203 pectinesterase homolog F4I1... 83 3e-16
>BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-like protein
- maize, partial (25%)
Length = 678
Score = 365 bits (938), Expect = e-101
Identities = 180/223 (80%), Positives = 198/223 (88%)
Frame = +1
Query: 285 GRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAM 344
GRY IYVKAGVYDEYIT+PKDAVNILMYGDGP +TIVTGRK+ AAGVKTMQTATFANTA+
Sbjct: 1 GRYTIYVKAGVYDEYITIPKDAVNILMYGDGPGKTIVTGRKNGAAGVKTMQTATFANTAL 180
Query: 345 GFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSG 404
GFIGKAMTFENTAGP GHQAVAFRNQGDMSALVGCHI+GYQD+LYVQ+NRQ+YRNC++SG
Sbjct: 181 GFIGKAMTFENTAGPAGHQAVAFRNQGDMSALVGCHILGYQDTLYVQTNRQFYRNCVISG 360
Query: 405 TVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFP 464
TVDFIFG+SATLIQ STIIVR P QFNTITADGS LNTGIVIQ CNI+P+AALFP
Sbjct: 361 TVDFIFGTSATLIQDSTIIVRMPSPNQFNTITADGSYVNKLNTGIVIQGCNIVPKAALFP 540
Query: 465 ERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQN 507
+RFTI+SYLGRPWK L KTVVMESTIG F + W G++N
Sbjct: 541 QRFTIKSYLGRPWKVLTKTVVMESTIG*F-YSSXWMDPLGKEN 666
>TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precursor (EC
3.1.1.11) (Pectin methylesterase 3) (PE 3)., partial
(93%)
Length = 2088
Score = 343 bits (881), Expect = 8e-95
Identities = 207/572 (36%), Positives = 309/572 (53%), Gaps = 13/572 (2%)
Frame = +2
Query: 5 VIISAVSLILVVGVAIGVVVAVRKNGED-------PEVQ-TQQRNLRIMCQNSQDQKLCH 56
+IIS++ L+ V+ A+ ++ + N E P + T +L+ +C+++Q C
Sbjct: 152 IIISSIILVAVIIAAVAGILIHKHNTESSSSPNSLPNTELTPATSLKAVCESTQYPNSCF 331
Query: 57 ETLSSVHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLM 116
++SS+ ++ DP+ S+K A D + K +S +E E K A+ C +++
Sbjct: 332 SSISSLPDSNTTDPEQLFKLSLKVAIDELSK---LSLTRFSEKATEPRVKKAIGVCDNVL 502
Query: 117 QFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDAND----GEKK 172
+LD L+ S + D +I D+ WLSA ++ C++ + N G
Sbjct: 503 ADSLDRLNDSMSTIVDGGKMLSPAKIRDVETWLSAALTDHDTCLDAVGEVNSTAARGVIP 682
Query: 173 IKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQGYP 232
E+ S + A+ ++ LS+ F+V RRLL +P
Sbjct: 683 EIERIMRNSTEFASNSLAIVSKVIRLLSN--------FEVSNHHRRLLGE--------FP 814
Query: 233 SWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVK 292
W+ +++R+LLA + + +P+AVVAKDGSGQ+KTI AL K + R+V+YVK
Sbjct: 815 EWLGTAERRLLATVVNET-----VPDAVVAKDGSGQYKTIGEALKLVKKKSLQRFVVYVK 979
Query: 293 AGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMT 352
GVY E I + K+ N+++YGDG T+V+G +++ G T +TATFA GFI K +
Sbjct: 980 KGVYVENIDLDKNTWNVMIYGDGMTETVVSGSRNYIDGTPTFETATFAVKGKGFIAKDIQ 1159
Query: 353 FENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGS 412
F NTAG HQAVA R+ D S C VGYQD+LY SNRQ+YR+C ++GT+DFIFG+
Sbjct: 1160FLNTAGASKHQAVAMRSGSDQSVFYRCSFVGYQDTLYAHSNRQFYRDCDITGTIDFIFGN 1339
Query: 413 SATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSY 472
+A + Q+ I+ R+P QFNTITA G N N+GIVIQ L + +Y
Sbjct: 1340AAAVFQNCKIMPRQPMSNQFNTITAQGKKDPNQNSGIVIQKSTF---TTLPGDNLIAPTY 1510
Query: 473 LGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGE-QNHNTCYYAEYANTGPGANVARRVKW 531
LGRPWK + T++M+S IG F+ P GW W + ++ YAEY NTGPGA+V RVKW
Sbjct: 1511LGRPWKDFSTTIIMKSEIGSFLKPVGWISWVANVEPPSSILYAEYQNTGPGADVPGRVKW 1690
Query: 532 KGYHGVISRAEANKFTAGIWLQAGPKSAAEWL 563
GY + +A KFT ++Q GP EWL
Sbjct: 1691AGYKPALGDEDAIKFTVDSFIQ-GP----EWL 1771
>TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Citrus
sinensis}, partial (85%)
Length = 1993
Score = 335 bits (859), Expect = 3e-92
Identities = 208/590 (35%), Positives = 306/590 (51%), Gaps = 20/590 (3%)
Frame = +1
Query: 4 KVIISAVSLILVVGVAIGVVVAVRKNGEDPEVQTQQRN-----------LRIMCQNSQDQ 52
K++IS + +L+V + +V KN + + + L+ C +
Sbjct: 103 KLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTTLYP 282
Query: 53 KLCHETLSSVHGAD--AADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALN 110
+LC +SS + K I+ S+ T V + E+L K+AL+
Sbjct: 283 ELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLTKREKIALH 462
Query: 111 DCKDLMQFALDSL-DLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGF--DDAN 167
DC + + LD L + V + + ++ D++ +S+ I+ + C++GF DDA+
Sbjct: 463 DCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSHDDAD 642
Query: 168 DGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDI----LQQFNLKFDVKPASRRLLNSE 223
+K+ ++ + V+ + + AL + ++D +Q N+ +R+LL E
Sbjct: 643 KEVRKVLQEGQIH----VEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNK-NRKLLEEE 807
Query: 224 VTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGN 283
V G+P WIS+ DR+LL + + + VVA DGSG FKT+ A+A+ P +
Sbjct: 808 NGV---GWPEWISAGDRRLLQG-------STVKADVVVAADGSGNFKTVSEAVAAAPLKS 957
Query: 284 KGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTA 343
RYVI +KAGVY E + VPK NI+ GDG TI+TG ++ G T +AT A
Sbjct: 958 SKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVG 1137
Query: 344 MGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVS 403
F+ + +TF+NTAGP HQAVA R D+SA C I+ YQD+LYV +NRQ++ NC +S
Sbjct: 1138GNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFIS 1317
Query: 404 GTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALF 463
GTVDFIFG+SA + Q+ I R+P GQ N +TA G N NTGIVIQ C I L
Sbjct: 1318GTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLE 1497
Query: 464 PERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGA 523
+ +YLGRPWK ++TV M+S+I D I P GW W G NT Y EY NTGPGA
Sbjct: 1498GVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGA 1677
Query: 524 NVARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
++RV WKG+ + S AEA T G ++ + WL P LG
Sbjct: 1678GTSKRVTWKGFKVITSAAEAQSSTPGNFI-----GGSSWLGSTGFPFSLG 1812
>TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11)
precursor - garden pea, complete
Length = 1860
Score = 322 bits (825), Expect = 2e-88
Identities = 187/522 (35%), Positives = 283/522 (53%), Gaps = 13/522 (2%)
Frame = +2
Query: 45 MCQNSQDQKLCHETLSSV-HGADAADPKAY----IAASVKAATDNVIKAFNMSERLTTEY 99
+C+++ D K C +S V G+ ++ K + + + + +T ++ KA + + +
Sbjct: 194 LCEHALDTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRV 373
Query: 100 GKENGAKMALNDCKDLMQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQAC 159
++ALNDC++LM ++D + S + NNI + HD WLS+V++ C
Sbjct: 374 NSPR-EEIALNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDA----HTWLSSVLTNHATC 538
Query: 160 MEGFDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRL 219
++G + G ++ + + L S + + L V A+
Sbjct: 539 LDGLE----GSSRVVMESDLHDLISRARSSLAVLVSVLP--------------PKANDGF 664
Query: 220 LNSEVTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASY 279
++ ++ D +PSW++S DR+LL +I N VVA+DGSG+FKT+ A+AS
Sbjct: 665 IDEKLNGD---FPSWVTSKDRRLLESSV-----GDIKANVVVAQDGSGKFKTVAQAVASA 820
Query: 280 PKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATF 339
P K RYVIYVK G Y E I + K N+++ GDG TI+TG +F G T ++AT
Sbjct: 821 PDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATV 1000
Query: 340 ANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRN 399
A GFI + + F+NTAGP HQAVA R D S + C I +QD+LY SNRQ+YR+
Sbjct: 1001AAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRD 1180
Query: 400 CLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPE 459
++GTVDFIFG++A + Q S + RKP Q N +TA G + N NT IQ C++IP
Sbjct: 1181SYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPS 1360
Query: 460 AALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNH--NTCYYAEYA 517
+ L P + +I++YLGRPWK ++TVV++S + I P GW W T YY EY
Sbjct: 1361SDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYM 1540
Query: 518 NTGPGANVARRVKWKGYHGVISRAEANKFT------AGIWLQ 553
N+G GA +RV W GYH + + AEA+KFT +WL+
Sbjct: 1541NSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWLK 1666
>NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1
Length = 1698
Score = 321 bits (822), Expect = 6e-88
Identities = 197/573 (34%), Positives = 309/573 (53%), Gaps = 6/573 (1%)
Frame = +1
Query: 7 ISAVSLILVVGVAIGVVVAVRKNGEDPE---VQTQQRNLRIMCQNSQDQKLCHETLSSVH 63
+S + L+ +VGV V V++ K G+ + + Q+N+ ++CQ+++ ++ CH+TL
Sbjct: 61 VSCILLVAMVGV---VAVSLTKGGDGEQKAHISNSQKNVDMLCQSTKFKETCHKTLEK-- 225
Query: 64 GADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLMQFALDSL 123
A ++ K I ++ A + + K N S L E ++ K A+ C +++ +A+D +
Sbjct: 226 -ASFSNMKNRIKGALGATEEELRKHINNSA-LYQELATDSMTKQAMEICNEVLDYAVDGI 399
Query: 124 DLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDAND--GEKKIKEQFHVQS 181
S + + + + D++ WL+ +S++Q C++GF + GE K
Sbjct: 400 HKSVGTLDQFDFHKLSEYAFDIKVWLTGTLSHQQTCLDGFVNTKTHAGETMAKV------ 561
Query: 182 LDSVQKVTAVALDIVTGLSDILQQFN-LKFDVKPASRRLLNSEVTVDDQGYPSWISSSDR 240
L + ++++ A+D++ +S IL+ F+ ++ V SRRLL+ D G PSW+S R
Sbjct: 562 LKTSMELSSNAIDMMDVVSRILKGFHPSQYGV---SRRLLS------DDGIPSWVSDGHR 714
Query: 241 KLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYI 300
LLA N+ NAVVA+DGSGQFKT+ AL + P N +VIYVKAGVY E +
Sbjct: 715 HLLAG-------GNVKANAVVAQDGSGQFKTLTDALKTVPPTNAAPFVIYVKAGVYKETV 873
Query: 301 TVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPD 360
V K+ + + GDGP +T TG ++A G+ T +TATF F+ K + FENTAG
Sbjct: 874 NVAKEMNYVTVIGDGPTKTKFTGSLNYADGINTYKTATFGVNGANFMAKDIGFENTAGTS 1053
Query: 361 GHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHS 420
QAVA R D + C + G+QD+L+V+S RQ+YR+C +SGT+DF+FG + + Q+
Sbjct: 1054KFQAVALRVTADQAIFHNCQMDGFQDTLFVESQRQFYRDCAISGTIDFVFGDAFGVFQNC 1233
Query: 421 TIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYL 480
+I R P KGQ +TA G D N + +V + E AL + SYLGRPWK
Sbjct: 1234KLICRVPAKGQKCLVTAGGRDKQNSASALVFLSSHFTGEPALTSVTPKL-SYLGRPWKLY 1410
Query: 481 AKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISR 540
+K V+M+STI P+G+ G +TC + EY N GPGA+ RVKW G + S
Sbjct: 1411SKVVIMDSTIDAMFAPEGYMPMVGGAFKDTCTFYEYNNKGPGADTNLRVKWHGVKVLTSN 1590
Query: 541 AEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
A + + + W+ VP+ LG
Sbjct: 1591VAAEYYPGKFFEIVNATARDTWIVKSGVPYSLG 1689
>TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, complete
Length = 1981
Score = 308 bits (789), Expect = 4e-84
Identities = 183/519 (35%), Positives = 276/519 (52%), Gaps = 10/519 (1%)
Frame = +3
Query: 45 MCQNSQDQKLCHETLSSV-HGADAADPKAY----IAASVKAATDNVIKAF---NMSERLT 96
+C+++ D C ++ V G+ + K + + + + +T ++ KA N+ +R
Sbjct: 201 VCEHAVDTNSCLTHVAEVVQGSTLDNTKDHKLSTLISLLTKSTTHIRKAMDTANVIKRRI 380
Query: 97 TEYGKENGAKMALNDCKDLMQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYR 156
+EN ALN C+ LM +++ + S + +N+ D D WLS+V++
Sbjct: 381 NSPREEN----ALNVCEKLMNLSMERVWDSVLTLTKDNM----DSQQDAHTWLSSVLTNH 536
Query: 157 QACMEGFDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPAS 216
C++G + G + + +Q L + + + L V D
Sbjct: 537 ATCLDGLE----GTSRAVMENDIQDLIARARSSLAVLVAVLPPKD--------------H 662
Query: 217 RRLLNSEVTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAAL 276
++ + D +PSW++S DR+LL ++ N VVAKDGSG+FKT+ A+
Sbjct: 663 DEFIDESLNGD---FPSWVTSKDRRLLESSV-----GDVKANVVVAKDGSGKFKTVAEAV 818
Query: 277 ASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQT 336
AS P RYVIYVK G+Y E + + N+++ GDG TI+TG ++ G T QT
Sbjct: 819 ASAPNKGTARYVIYVKKGIYKENVEIASSKTNVMLLGDGMDATIITGSLNYVDGTGTFQT 998
Query: 337 ATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQY 396
AT A FI + + F+NTAGP HQAVA R D S + C I +QD+LY +NRQ+
Sbjct: 999 ATVAAVGDWFIAQDIGFQNTAGPQKHQAVALRVGSDRSVINRCKIDAFQDTLYAHTNRQF 1178
Query: 397 YRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNI 456
YR+ ++GT+DFIFG +A ++Q ++ RKP Q N +TA G N NT IQ C++
Sbjct: 1179YRDSFITGTIDFIFGDAAVVLQKCKLVARKPMANQNNMVTAQGRIDPNQNTATSIQQCDV 1358
Query: 457 IPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNH--NTCYYA 514
IP L P ++++YLGRPWK ++TVVM+S +G I P GW W T YY
Sbjct: 1359IPSTDLKPVIGSVKTYLGRPWKKYSRTVVMQSLLGAHIDPTGWAEWDAASKDFLQTLYYG 1538
Query: 515 EYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQ 553
EY N+GPGA ++RVKW GYH +I+ AEANKFT +Q
Sbjct: 1539EYMNSGPGAGTSKRVKWPGYH-IINTAEANKFTVAQLIQ 1652
>TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, partial (98%)
Length = 2029
Score = 301 bits (770), Expect = 6e-82
Identities = 176/517 (34%), Positives = 272/517 (52%), Gaps = 7/517 (1%)
Frame = +2
Query: 44 IMCQNSQDQKLCHETLSSVHGADAA----DPKAYIAASVKA-ATDNVIKAFNMSERLTTE 98
+ C+++ D K C +S V D ++ S+ +T ++ A + + +
Sbjct: 206 VYCEHAVDTKSCLAHVSEVSHVPTLVTTKDQNLHVLLSLLTKSTTHIQNAMDTASVIKRR 385
Query: 99 YGKENGAKMALNDCKDLMQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQA 158
++AL+DC+ LM +++ + + + NNI D D WLS+V++
Sbjct: 386 INSPR-EEIALSDCEQLMDLSMNRIWDTMLKLTKNNI----DSQQDAHTWLSSVLTNHAT 550
Query: 159 CMEGFDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRR 218
C++G + G ++ + +Q D+++ L F + F K +
Sbjct: 551 CLDGLE----GSSRVVMENDLQ-------------DLISRARSSLAVFLVVFPQKDRDQF 679
Query: 219 LLNSEVTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALAS 278
+ E + + +PSW++S DR+LL +I N VVA+DGSG+FKT+ A+AS
Sbjct: 680 I--DETLIGE--FPSWVTSKDRRLLETAV-----GDIKANVVVAQDGSGKFKTVAEAVAS 832
Query: 279 YPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTAT 338
P K +YVIYVK G Y E + + N+++ GDG TI+TG +F G T +++T
Sbjct: 833 APDNGKTKYVIYVKKGTYKENVEIGSKKTNVMLVGDGMDATIITGNLNFIDGTTTFKSST 1012
Query: 339 FANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYR 398
A GFI + + F+N AG HQAVA R D S + C I +QD+LY SNRQ+YR
Sbjct: 1013VAAVGDGFIAQDIWFQNMAGAAKHQAVALRVGSDQSVINRCRIDAFQDTLYAHSNRQFYR 1192
Query: 399 NCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIP 458
+ +++GT+DFIFG++A + Q ++ RKP Q N TA G + NTG IQ C++ P
Sbjct: 1193DSVITGTIDFIFGNAAVVFQKCKLVARKPMANQNNMFTAQGREDPGQNTGTSIQQCDLTP 1372
Query: 459 EAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNH--NTCYYAEY 516
+ L P +I+++LGRPWK ++TVVM+S + I P GW W T YY EY
Sbjct: 1373SSDLKPVVGSIKTFLGRPWKKYSRTVVMQSFLDSHIDPTGWAEWDAASKDFLQTLYYGEY 1552
Query: 517 ANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQ 553
N GPGA A+RV W GYH + + AEA+KFT +Q
Sbjct: 1553LNNGPGAGTAKRVTWPGYHVINTAAEASKFTVAQLIQ 1663
>TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 {Arabidopsis
thaliana}, partial (79%)
Length = 2442
Score = 300 bits (769), Expect = 8e-82
Identities = 177/470 (37%), Positives = 250/470 (52%), Gaps = 2/470 (0%)
Frame = +3
Query: 106 KMALNDCKDLMQFALDSLDLS--TKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGF 163
K A DC +L F + L + TKC + + WLS ++ + C GF
Sbjct: 843 KAAWADCLELYDFTVQKLSQTKYTKCTQ-----------YESQTWLSTALTNLETCKNGF 989
Query: 164 DDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSE 223
D VT L +++ L L + P +
Sbjct: 990 YDLG--------------------VTNYVLPLLSNNVTKLLSNTLSLNKVPYQQPSYKD- 1106
Query: 224 VTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGN 283
G+P+W+ DRKLL A N VVAKDGSG++ T++AA + P G+
Sbjct: 1107 ------GFPTWVKPGDRKLLQTSS-----AASKANVVVAKDGSGKYTTVKAATDAAPSGS 1253
Query: 284 KGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTA 343
GRYVIYVKAGVY+E + + A N+++ GDG +TI+TG KS G T ++AT A T
Sbjct: 1254 -GRYVIYVKAGVYNEQVEIK--AKNVMLVGDGIGKTIITGSKSVGGGTTTFRSATVAATG 1424
Query: 344 MGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVS 403
GFI + +TF NTAG HQAVAFR+ D+S C GYQD+LYV S RQ+YR C +
Sbjct: 1425 DGFIAQDITFRNTAGAANHQAVAFRSGSDLSVFYKCSFEGYQDTLYVHSERQFYRECNIY 1604
Query: 404 GTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALF 463
GTVDFIFG++A ++Q+ I R P + T+TA G N NTGI+I + + ++ L
Sbjct: 1605 GTVDFIFGNAAVVLQNCNIFARNP-PAKTITVTAQGRTDPNQNTGIIIHNSRVSAQSDLN 1781
Query: 464 PERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGA 523
P +++SYLGRPW+ ++TV M++ + FI+P GW W G +T YYAEYANTG G+
Sbjct: 1782 PS--SVKSYLGRPWQKYSRTVFMKTVLDGFINPAGWLPWDGNFALDTLYYAEYANTGSGS 1955
Query: 524 NVARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
+ + RV WKGYH + S ++A+ FT G ++ + W+ VP G
Sbjct: 1956 STSNRVTWKGYHVLTSASQASPFTVGNFI-----AGNSWIGNTGVPFTSG 2090
>TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylesterase isoform
alpha {Vigna radiata}, partial (94%)
Length = 1655
Score = 278 bits (710), Expect = 5e-75
Identities = 145/321 (45%), Positives = 194/321 (60%), Gaps = 1/321 (0%)
Frame = +2
Query: 234 WISSSDRKLL-AKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVK 292
W+S DRKLL A + + + + VVAKDG+G F TI A+A+ P + R+VI++K
Sbjct: 41 WVSPKDRKLLQAAVNQTKF------DLVVAKDGTGNFATIGEAIAAAPNSSATRFVIHIK 202
Query: 293 AGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMT 352
AG Y E + V K N+++ GDG +T+V ++ G T Q++TFA FI K +T
Sbjct: 203 AGAYFENVEVIKKKTNLMLVGDGIGQTVVKASRNVVDGWTTFQSSTFAVVGDKFIAKGIT 382
Query: 353 FENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGS 412
FEN+AGP HQAVA RN D SA C V YQD+LYV S RQ+YR C V GTVDFIFG+
Sbjct: 383 FENSAGPSKHQAVAVRNGADFSAFYQCSFVAYQDTLYVHSLRQFYRECDVYGTVDFIFGN 562
Query: 413 SATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSY 472
+A + Q+ + RKP Q N TA G + N NTGI I +C I A L P + T +SY
Sbjct: 563 AAVVFQNCNLYARKPDPKQKNLFTAQGREDPNQNTGISILNCKIAAAADLIPVQSTFKSY 742
Query: 473 LGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWK 532
LGRPWK ++TV + S I + I P GW W G +T +Y EY N GPG+N + RV W
Sbjct: 743 LGRPWKKYSRTVYLNSFIDNLIDPPGWLEWNGTFALDTLFYGEYKNRGPGSNTSARVTWP 922
Query: 533 GYHGVISRAEANKFTAGIWLQ 553
GY + + EA++FT ++Q
Sbjct: 923 GYKVITNATEASQFTVRQFIQ 985
>TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1929
Score = 270 bits (690), Expect = 1e-72
Identities = 179/544 (32%), Positives = 265/544 (47%), Gaps = 23/544 (4%)
Frame = +3
Query: 46 CQNSQDQKLCHETL----SSVHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGK 101
C+++ D C L ++V+ K ++ S K + D + K L+T
Sbjct: 138 CKSTPDPTYCKSILPPQNANVYDYGRFSVKKSLSQSRKFS-DLINKYLQRRSTLSTT--- 305
Query: 102 ENGAKMALNDCKDLMQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACME 161
A AL DC+ L D L S + V+ Q +++ LSA+++ +Q C++
Sbjct: 306 ---ALRALQDCQSLSDLNFDFLSSSFQTVNKTTKFLPSLQGENIQTLLSAILTNQQTCLD 476
Query: 162 GFDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLN 221
G D + S S + + L T L + F K V P + +
Sbjct: 477 GLKDTS-------------SAWSFRNGLTIPLSNDTKLYSVSLAFFTKGWVNPKTNKTSF 617
Query: 222 SEVTVDDQGY-----PSWISSSDRKLLAKMQRKNWRAN--------IMPNAVVAKDGSGQ 268
++G+ P ++S R + + R+ + + V++DGSG
Sbjct: 618 PNSKHSNKGFKNGRLPLKMTSKTRAIYESVSRRKLLQSNQVGEDVVVRDIVTVSQDGSGN 797
Query: 269 FKTIQAALASYPK---GNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRK 325
F TI A+A+ P + G ++IYV AGVY+EYIT+ K ++M GDG +TI+TG
Sbjct: 798 FTTINDAIAAAPNKSVSSDGYFLIYVTAGVYEEYITIDKKKTYLMMIGDGINKTIITGNH 977
Query: 326 SFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQ 385
S G T + TFA GF+G MT NTAG HQAVA RN D+S C GYQ
Sbjct: 978 SVVDGWTTFGSPTFAVVGQGFVGVNMTIRNTAGAVKHQAVALRNGADLSTFYSCSFEGYQ 1157
Query: 386 DSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNL 445
D+LY S RQ+YR C + GTVDFIFG++ + Q+ + R P GQFN ITA G N
Sbjct: 1158DTLYTHSLRQFYRECDIYGTVDFIFGNAKVVFQNCNLYPRLPMSGQFNAITAQGRTDPNQ 1337
Query: 446 NTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGE 505
+TG I +C I L + +YLGRPWK ++TV M++ + + I+ GW W GE
Sbjct: 1338DTGTSIHNCTIKATDDLAASNGAVSTYLGRPWKEYSRTVYMQTFMDNVINVAGWRAWDGE 1517
Query: 506 QNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAG---PKSAAEW 562
+T YYAE+ N+GPG++ RV W+GYH VI+ +A FT +L P++ +
Sbjct: 1518FALSTLYYAEFNNSGPGSSTDGRVTWQGYH-VINATDAANFTVANFLLGDDWLPQTGVSY 1694
Query: 563 LNGL 566
N L
Sbjct: 1695TNSL 1706
>TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectinesterase
{Arabidopsis thaliana}, partial (59%)
Length = 1963
Score = 265 bits (678), Expect = 3e-71
Identities = 179/525 (34%), Positives = 265/525 (50%), Gaps = 23/525 (4%)
Frame = +2
Query: 46 CQNSQDQKLCHETLSSVHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGA 105
C+ + KLC LS++ + + DP Y S+K +K E++ ++ + +
Sbjct: 155 CKTTLYPKLCRSMLSAIRSSPS-DPYNYGKFSIKQN----LKVARKLEKVFIDFLNRHQS 319
Query: 106 KMALN--------DCKDLMQFALDSLDLST---KCVHDNNIQAVHDQIADMRNWLSAVIS 154
+LN DCKDL +D L+ + K ++ + + + + ++LSAV +
Sbjct: 320 SSSLNHEEVGALVDCKDLNSLNVDYLESISDELKSASSSSSSSDTELVDKIESYLSAVAT 499
Query: 155 YRQACMEGFDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVT-GLSDILQQF------- 206
C +G + I V D+ Q +V+L +VT LS +++
Sbjct: 500 NHYTCYDGLVVT---KSNIANALAVPLKDATQ-FYSVSLGLVTEALSKNMKRNKTRKHGL 667
Query: 207 -NLKFDVKPASRRLLNSEVTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDG 265
N F V+ +L+ T S +S R + ++ + +V+ G
Sbjct: 668 PNKSFKVRQPLEKLIKLLRTKYSCQKTSSNCTSTRTERILKESESHGILLNDFVLVSPYG 847
Query: 266 SGQFKTIQAALASYPKGNK---GRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVT 322
+I A+A+ P K G Y+IYV+ G Y+EY+ VPK NIL+ GDG TI+T
Sbjct: 848 IANHTSIGDAIAAAPNNTKPEDGYYLIYVREGYYEEYVIVPKHKNNILLVGDGINNTIIT 1027
Query: 323 GRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIV 382
G S G T ++TFA + FI +TF NTAGP+ HQAVA RN D+S C
Sbjct: 1028GNHSVIDGWTTFNSSTFAVSGERFIAVDITFRNTAGPEKHQAVAVRNNADLSTFYRCSFE 1207
Query: 383 GYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDT 442
GYQD+LYV S RQ+YR+C + GTVDFIFG++A + Q+ I RKP Q N +TA G
Sbjct: 1208GYQDTLYVHSVRQFYRDCKIYGTVDFIFGNAAVVFQNCNIYARKPLPNQKNAVTAQGRTD 1387
Query: 443 MNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIW 502
N NTGI IQ+C I L + + SYLGRPWK ++TV M+S IGDF+ P GW W
Sbjct: 1388PNQNTGISIQNCTIDAAQDLANDLNSTMSYLGRPWKIYSRTVYMQSYIGDFVQPSGWLEW 1567
Query: 503 QGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFT 547
G +T +Y E+ N GPG+ RV+W G H +++ +A FT
Sbjct: 1568NGTVGLDTIFYGEFNNYGPGSVTNNRVQWPG-HFLLNDTQAWNFT 1699
>TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectinesterase
{Arabidopsis thaliana}, partial (34%)
Length = 691
Score = 194 bits (494), Expect = 6e-50
Identities = 104/216 (48%), Positives = 133/216 (61%)
Frame = +3
Query: 232 PSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYV 291
P+W+ DRKLL K +A+I VVAKDGSG++KTI AAL P + R VIYV
Sbjct: 54 PNWLHHKDRKLLQKDSDLKKKADI----VVAKDGSGKYKTISAALKHVPNKSDKRTVIYV 221
Query: 292 KAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAM 351
K G+Y E + V K N+++ GDG TIV+G +F G T TATFA FI + +
Sbjct: 222 KKGIYYENVRVEKTKWNVMIIGDGMNVTIVSGSLNFIDGTPTFSTATFAVFGRNFIARDI 401
Query: 352 TFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFG 411
F+NTAGP HQAVA D + C + YQD+LY SNRQ+YR C + GTVDFIFG
Sbjct: 402 GFKNTAGPQKHQAVALMTSADQAVFYKCSMDAYQDTLYAHSNRQFYRECNIYGTVDFIFG 581
Query: 412 SSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNT 447
+SA ++Q+ I+ R+P GQ NTITA G N+NT
Sbjct: 582 NSAVVLQNCNILPRQPMPGQQNTITAQGKTDPNMNT 689
>BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like protein -
Arabidopsis thaliana, partial (36%)
Length = 658
Score = 167 bits (424), Expect = 8e-42
Identities = 91/205 (44%), Positives = 125/205 (60%), Gaps = 9/205 (4%)
Frame = +1
Query: 283 NKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAA-GVKTMQTATFAN 341
++ R+VIY+K GVY+E + VP N++ GDG +T++TG + G+ T +AT A
Sbjct: 37 DRKRFVIYIKEGVYEETVRVPLKKRNVVFLGDGIGKTVITGSANVGQPGMTTYNSATVAV 216
Query: 342 TAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCL 401
GF+ K +T ENTAGPD HQAVAFR D+S + C +G QD+LY S RQ+Y++C
Sbjct: 217 LGDGFMAKDLTIENTAGPDAHQAVAFRLDSDLSVIENCEFLGNQDTLYAHSLRQFYKSCR 396
Query: 402 VSGTVDFIFGSSATLIQHSTIIVR----KPGKGQFNTITADGSDTMNLNTGIVIQDCNI- 456
+ G VDFIFG+SA + Q I+VR KP KG+ N ITA G +TG V Q+C I
Sbjct: 397 IVGNVDFIFGNSAAIFQDCQILVRPRQLKPEKGENNAITAHGRTDPAQSTGFVFQNCLIN 576
Query: 457 --IPEAALFPERFTI-RSYLGRPWK 478
AL+ + ++YLGRPWK
Sbjct: 577 GTEDYMALYHSNPKVHKNYLGRPWK 651
>TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor (EC
3.1.1.11) (Pectin methylesterase) (PE) (P65). [Alfalfa],
partial (53%)
Length = 732
Score = 157 bits (398), Expect = 8e-39
Identities = 84/211 (39%), Positives = 119/211 (55%), Gaps = 3/211 (1%)
Frame = +2
Query: 342 TAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCL 401
T+ F + FEN+AG HQAVA R D + C + GYQD+LY QS RQ+YR+C
Sbjct: 74 TSAHFTALNVGFENSAGAAKHQAVALRVTADKALFYNCQMNGYQDTLYTQSKRQFYRDCT 253
Query: 402 VSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAA 461
++GT+DF+FG + + Q+ +IVRKP Q +TA G ++ + +V Q+C+ E
Sbjct: 254 ITGTIDFVFGDAVGVFQNCKLIVRKPMANQQCMVTAGGRTKVDSVSALVFQNCHFTGE-- 427
Query: 462 LFPERFTIR---SYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYAN 518
PE T++ +YLGRPW+ +K V+++S I P+G+ W G TC Y EY N
Sbjct: 428 --PEVLTMQPKIAYLGRPWRNFSKVVIVDSLIDGLFVPEGYMPWMGNLFKETCTYLEYNN 601
Query: 519 TGPGANVARRVKWKGYHGVISRAEANKFTAG 549
G GA +VKW G IS EA K+ G
Sbjct: 602 KGAGAATNLKVKWPGVK-TISAGEAAKYYPG 691
>BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precursor (EC
3.1.1.11) (Pectin methylesterase) (PE). [Peach] {Prunus
persica}, partial (36%)
Length = 589
Score = 153 bits (386), Expect = 2e-37
Identities = 71/162 (43%), Positives = 107/162 (65%)
Frame = +2
Query: 265 GSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGR 324
GSG + + A+++ P+ + RYVIYVK GVY E + + K NI++ G+G TI++G
Sbjct: 2 GSGDYAKVMDAVSAAPESSMKRYVIYVKKGVYVENVEIKKKKWNIMLIGEGMDATIISGS 181
Query: 325 KSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGY 384
+++ G T ++ATFA + GFI + ++F+NTAG + HQAVA R+ D+S C I GY
Sbjct: 182 RNYVDGSTTFRSATFAVSGRGFIARDISFQNTAGAEKHQAVALRSDSDLSVFYRCGIFGY 361
Query: 385 QDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRK 426
QDSLY + RQ+YR C +SGTVDFIFG + + Q+ I+ ++
Sbjct: 362 QDSLYTHTMRQFYRECKISGTVDFIFGDATAVFQNCQILAKE 487
>TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pectin
methylesterase {Arabidopsis thaliana}, partial (33%)
Length = 1289
Score = 109 bits (273), Expect(2) = 7e-31
Identities = 87/280 (31%), Positives = 125/280 (44%), Gaps = 24/280 (8%)
Frame = +3
Query: 222 SEVTVDDQGYPSW-ISSSDRKLLAKMQRKNWRANIM----------PNAV---VAKDGSG 267
SE+ D + SW + + RK + + + +N N+ N V V++DGS
Sbjct: 84 SEMESDFDKWISWNVKNYQRKTIMEKRYRNVSGNVQGLDPKLKKAESNKVRLKVSQDGSA 263
Query: 268 QFKTIQAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRK-- 325
QFK+I AL S N R +I + G Y E I VPK I GD +TG
Sbjct: 264 QFKSITEALNSIQPYNIRRVIISIAPGYYREKIVVPKTLPFITFLGDVRDPPTITGNDTQ 443
Query: 326 ----SFAAGVKTMQTATFANTAMGFIGKAMTFENTA----GPDGHQAVAFRNQGDMSALV 377
S A ++T +AT A A F+ + FENTA G QAVA R G+ +A
Sbjct: 444 SVTGSDGAQLRTFNSATVAVNASYFMAININFENTASFPIGSKVEQAVAVRITGNKTAFY 623
Query: 378 GCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITA 437
C G QD+LY Y+ NC + G+VDFI G +L + TI + +ITA
Sbjct: 624 NCTFSGVQDTLYDHKGLHYFNNCTIKGSVDFICGHGKSLYEGCTI---RSIANNMTSITA 794
Query: 438 DGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPW 477
+ ++G ++ +I + +YLGRPW
Sbjct: 795 QSGSNPSYDSGFSFKNSMVIGDGP---------TYLGRPW 887
Score = 42.7 bits (99), Expect(2) = 7e-31
Identities = 16/36 (44%), Positives = 20/36 (55%)
Frame = +1
Query: 496 PDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKW 531
P GW W + + YY EY +GPG+N A RV W
Sbjct: 943 PKGWEDWNDTKRYMNAYYGEYKCSGPGSNTAGRVPW 1050
>AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (25%)
Length = 495
Score = 127 bits (319), Expect = 1e-29
Identities = 63/145 (43%), Positives = 90/145 (61%), Gaps = 3/145 (2%)
Frame = +1
Query: 263 KDGSGQFKTIQAALASYPKG---NKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPART 319
+DGSG F TI A+A+ P + G ++I++ GVY+EY+++ K ++M G+G +T
Sbjct: 61 QDGSGNFTTINDAIAAAPNNTVASDGYFLIFITEGVYEEYVSIDKKKKYLMMIGEGINQT 240
Query: 320 IVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGC 379
I+TG ++ A G T +ATFA A GF+ +TF NTAG +QAVA R+ DMS C
Sbjct: 241 IITGNRNVADGFTTFNSATFAVVAQGFVAVNITFRNTAGAAKNQAVALRSGADMSTFYSC 420
Query: 380 HIVGYQDSLYVQSNRQYYRNCLVSG 404
GYQD+LY S RQ+YR C + G
Sbjct: 421 SFEGYQDTLYTHSLRQFYRECDIYG 495
>TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE8B
precursor (EC 3.1.1.11) (Pectin methylesterase) (PE).
[Peach] {Prunus persica}, partial (34%)
Length = 1415
Score = 119 bits (298), Expect = 3e-27
Identities = 89/299 (29%), Positives = 143/299 (47%), Gaps = 18/299 (6%)
Frame = +1
Query: 59 LSSVHGADAADPKAYIAASVKAAT-------DNVI----KAFNMSERLTTEYGKENGAKM 107
LS H A+ P ++ +S++ +T + VI K ++ R T +G +
Sbjct: 148 LSLPHSTFASSPNDFVGSSLRVSTAKFANSAEEVITVLQKVISILSRFTNVFGHSRTSN- 324
Query: 108 ALNDCKDLMQFALDSLDLSTKCVH-----DNNIQAVHDQIADMRNWLSAVISYRQACMEG 162
A++DC DL+ +LD L+ S DN+ ++ D+R WLSAV+ Y C+EG
Sbjct: 325 AVSDCLDLLDMSLDQLNQSISAAQKPKEKDNSTGKLN---CDLRTWLSAVLVYPDTCIEG 495
Query: 163 FDDANDGEKKIKEQFHVQSLDSVQKVTAVAL-DIVTGLSDILQQFNLKFDVKPASRRLLN 221
E I + LD V + A L ++V+G D
Sbjct: 496 L------EGSIVKGLISSGLDHVMSLVANLLGEVVSGNDD-------------------- 597
Query: 222 SEVTVDDQGYPSWISSSDRKLLAKMQRKNWRAN-IMPNAVVAKDGSGQFKTIQAALASYP 280
++ + +PSWI D KLL +AN + +AVVA DGSG + + A+++ P
Sbjct: 598 -QLATNKDRFPSWIRDEDTKLL--------QANGVTADAVVAADGSGDYAKVMDAVSAAP 750
Query: 281 KGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATF 339
+ + RYVIYVK GVY E + + K NI++ G+G TI++G +++ G T ++ATF
Sbjct: 751 ESSMKRYVIYVKKGVYVENVEIKKKKWNIMLIGEGMDATIISGSRNYVDGSTTFRSATF 927
>TC93392 similar to PIR|T49241|T49241 pectinesterase-like protein -
Arabidopsis thaliana, partial (18%)
Length = 467
Score = 102 bits (254), Expect = 4e-22
Identities = 47/100 (47%), Positives = 62/100 (62%), Gaps = 3/100 (3%)
Frame = +2
Query: 472 YLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKW 531
YLGRPWK ++T+ M+S + D I P+GW W G NT YYAEY N+GPGA VA RVKW
Sbjct: 2 YLGRPWKTYSRTIFMQSYMSDAIRPEGWLEWNGNFALNTLYYAEYMNSGPGAGVANRVKW 181
Query: 532 KGYHGVISRAEANKFTAGIWLQAG---PKSAAEWLNGLHV 568
GYH + +EA KFT +++ P + + +GL V
Sbjct: 182 SGYHVLNDSSEATKFTVAQFIEGNLGLPSTGVTYTSGLKV 301
>TC92167 similar to PIR|T05203|T05203 pectinesterase homolog F4I10.160 -
Arabidopsis thaliana, partial (11%)
Length = 652
Score = 83.2 bits (204), Expect = 3e-16
Identities = 55/147 (37%), Positives = 75/147 (50%), Gaps = 5/147 (3%)
Frame = +1
Query: 353 FENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYY----RNCLVSGTVDF 408
F NTAGP+GHQAVA R Q D + C G+QD+LY ++RQ++ N DF
Sbjct: 220 FRNTAGPEGHQAVAARVQADRAVFANCRFEGFQDTLYTVAHRQFFP*LHHN---REQFDF 390
Query: 409 IFGSSATLIQHSTIIVRKPGKGQFNTITADGSD-TMNLNTGIVIQDCNIIPEAALFPERF 467
IFG + L + + K T D T N + + ++ F +
Sbjct: 391 IFGDPSPLYSKTASWLLKNQVSAKVTR*QHKEDWTTNKIQQLFYINVHLKLMTHWFLLKA 570
Query: 468 TIRSYLGRPWKYLAKTVVMESTIGDFI 494
T++SYLGR WK ++TVVMES IGDFI
Sbjct: 571 TVKSYLGRAWKQFSRTVVMESDIGDFI 651
Score = 58.5 bits (140), Expect(2) = 5e-11
Identities = 47/180 (26%), Positives = 79/180 (43%), Gaps = 12/180 (6%)
Frame = +3
Query: 297 DEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENT 356
DE +T+ +I +YGDG ++I+TG K+F GV + TA+ GF+G A +
Sbjct: 51 DEVVTITDKMKDITIYGDGSQKSIITGSKNFRDGVTNINTASLVVLGEGFLGLASGIQKH 230
Query: 357 AGPDGHQAVAFRNQGDMSALVGCHI-----------VGYQDSLYVQSNRQYYRNCLVSGT 405
+ + + + + C + G L V ++ Q GT
Sbjct: 231 SW--S*RTSSSSS*SPSRSCCVCQLPF*RLPRHAIHSGT*TILPVAAS*Q--------GT 380
Query: 406 VDF-IFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFP 464
+ F ++ + Q+ ++V+KP GQ N +TA G NT IV+ C + + AL P
Sbjct: 381 IRFHLW*PQPVIFQNCILVVKKPSVGQSNAVTAQGRLDNKQNTAIVLHKCTLKADDALVP 560
Score = 26.9 bits (58), Expect(2) = 5e-11
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +2
Query: 285 GRYVIYVKAGVY 296
GRYVIYVK GVY
Sbjct: 14 GRYVIYVKEGVY 49
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,182,489
Number of Sequences: 36976
Number of extensions: 205159
Number of successful extensions: 909
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 859
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 864
length of query: 576
length of database: 9,014,727
effective HSP length: 101
effective length of query: 475
effective length of database: 5,280,151
effective search space: 2508071725
effective search space used: 2508071725
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144563.2


