

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144541.17 - phase: 0 /pseudo
(375 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
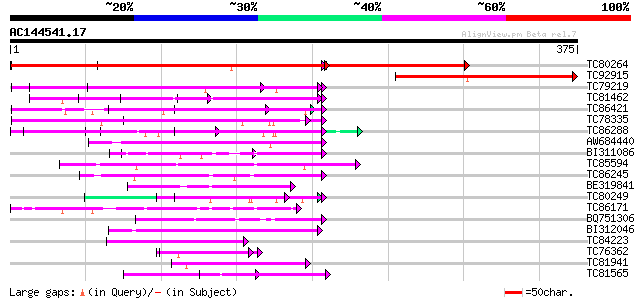
Score E
Sequences producing significant alignments: (bits) Value
TC80264 similar to GP|21689699|gb|AAM67471.1 putative F-box fami... 427 e-170
TC92915 similar to PIR|A27288|A27288 vicilin precursor - fava be... 166 1e-41
TC79219 similar to GP|15810000|gb|AAL06927.1 AT5g23340/MKD15_20 ... 97 8e-21
TC81462 similar to GP|21536497|gb|AAM60829.1 F-box protein famil... 87 8e-18
TC86421 similar to GP|2407790|gb|AAB70660.1| grr1 {Glycine max},... 84 1e-16
TC78335 similar to GP|16604366|gb|AAL24189.1 At1g21410/F24J8_17 ... 77 1e-14
TC86288 similar to GP|2407790|gb|AAB70660.1| grr1 {Glycine max},... 70 1e-12
AW684440 weakly similar to PIR|A84649|A84 probable glucose regul... 63 2e-10
BI311086 53 2e-07
TC85594 similar to PIR|D96512|D96512 hypothetical protein F2G19.... 53 2e-07
TC86245 weakly similar to GP|9279671|dbj|BAB01228.1 transport in... 52 3e-07
BE319841 weakly similar to GP|21592320|gb| unknown {Arabidopsis ... 52 5e-07
TC80249 weakly similar to GP|10716947|gb|AAG21976.1 SKP1 interac... 50 1e-06
TC86171 weakly similar to GP|13507547|gb|AAK28636.1 unknown prot... 49 2e-06
BQ751306 similar to GP|18376293|em related to protein GRR1 {Neur... 48 7e-06
BI312046 similar to GP|17979057|gb unknown protein {Arabidopsis ... 47 2e-05
TC84223 weakly similar to GP|18252175|gb|AAL61920.1 unknown prot... 45 3e-05
TC76362 similar to GP|10716949|gb|AAG21977.1 SKP1 interacting pa... 45 5e-05
TC81941 similar to GP|15420162|gb|AAK97303.1 F-box containing pr... 44 8e-05
TC81565 similar to GP|6041850|gb|AAF02159.1| unknown protein {Ar... 43 2e-04
>TC80264 similar to GP|21689699|gb|AAM67471.1 putative F-box family protein
AtFBL3 {Arabidopsis thaliana}, partial (4%)
Length = 1305
Score = 427 bits (1097), Expect(2) = e-170
Identities = 211/211 (100%), Positives = 211/211 (100%)
Frame = +3
Query: 1 AIDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVR 60
AIDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVR
Sbjct: 276 AIDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVR 455
Query: 61 VSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTI 120
VSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTI
Sbjct: 456 VSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTI 635
Query: 121 ANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVR 180
ANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVR
Sbjct: 636 ANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVR 815
Query: 181 LKLGLCTNISDIGLAHIACNCPKLTELDLYR 211
LKLGLCTNISDIGLAHIACNCPKLTELDLYR
Sbjct: 816 LKLGLCTNISDIGLAHIACNCPKLTELDLYR 908
Score = 189 bits (481), Expect(2) = e-170
Identities = 94/95 (98%), Positives = 95/95 (99%)
Frame = +2
Query: 210 YRR*WASSTDNRMQQVGHAQPSILQ*DYRRRVEVYQQSR*TL*F*VAWAFKHHEHWYKSS 269
YRR*WASSTDNRMQQVGHAQPSILQ*DYRRRVEVYQQSR*TL*F*VAWAFKHHEHWYKSS
Sbjct: 917 YRR*WASSTDNRMQQVGHAQPSILQ*DYRRRVEVYQQSR*TL*F*VAWAFKHHEHWYKSS 1096
Query: 270 CSQLQEIG*FRFEALRKT**YGFSGTCFLFTKPTA 304
CSQLQEIG*F+FEALRKT**YGFSGTCFLFTKPTA
Sbjct: 1097 CSQLQEIG*FKFEALRKT**YGFSGTCFLFTKPTA 1201
Score = 93.2 bits (230), Expect = 1e-19
Identities = 59/209 (28%), Positives = 107/209 (50%), Gaps = 1/209 (0%)
Frame = +3
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGV-R 60
+ +S+C V+ G++ V+ G L ++ C A ++ + +L+ ++++
Sbjct: 510 LGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDM 686
Query: 61 VSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTI 120
V++ L IGS+C L EL L+ C GV ++ + + C L L L C ++D ++ I
Sbjct: 687 VTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHI 866
Query: 121 ANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVR 180
A +CP L L L C + + GL + + C L L+L C+ + D LK +S +L
Sbjct: 867 ACNCPKLTELDLYRCVRIGDDGLAALTTGCNKLAMLNLAYCNRITDAGLKCISNLGELSD 1046
Query: 181 LKLGLCTNISDIGLAHIACNCPKLTELDL 209
+L +NI+ IG+ +A +C +L L+L
Sbjct: 1047FELRGLSNITSIGIKAVAVSCKRLANLNL 1133
Score = 56.2 bits (134), Expect = 2e-08
Identities = 45/202 (22%), Positives = 72/202 (35%), Gaps = 51/202 (25%)
Frame = +3
Query: 59 VRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAIS 118
+ +SD + ++ C L L +S + VTN + + L + C V DA +
Sbjct: 69 LEISDLGIDLLSKKCFDLNFLDVSY-LKVTNESLRSIASLLKLEVFIMVGCYLVDDAGLQ 245
Query: 119 TIANSCPNLACLKLESCDMVTEIGLYQ--------------------------------- 145
+ CP L + + C+ V+ GL
Sbjct: 246 FLEKGCPLLKAIDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKH 425
Query: 146 ------------------IGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVRLKLGLCT 187
IGS+C L EL L+ C GV ++ + + C L L L C
Sbjct: 426 LSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCR 605
Query: 188 NISDIGLAHIACNCPKLTELDL 209
++D ++ IA +CP L L L
Sbjct: 606 FVTDAAISTIANSCPNLACLKL 671
Score = 31.6 bits (70), Expect = 0.52
Identities = 19/45 (42%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Frame = +3
Query: 166 DIAL-KYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
DI L K CSKL +L L C ISD+G+ ++ C L LD+
Sbjct: 3 DIGLAKIAVGCSKLEKLSLKWCLEISDLGIDLLSKKCFDLNFLDV 137
>TC92915 similar to PIR|A27288|A27288 vicilin precursor - fava bean, partial
(4%)
Length = 862
Score = 166 bits (420), Expect = 1e-41
Identities = 90/129 (69%), Positives = 94/129 (72%), Gaps = 9/129 (6%)
Frame = -1
Query: 256 AWAFKHHEHWYKSSCSQLQEIG*FRFEALRKT**YGFSGTCFLFTK---------PTADH 306
AWAFKHHEH Y +Q * E++RK SGT F +DH
Sbjct: 847 AWAFKHHEHLY*KQLQSVQRDC*I*IESMRKNLMIRVSGTAFYSQNLLQINMSYCNVSDH 668
Query: 307 VLWLLMSNLKRLQDAKLVYLVNVTIQGLELALISCCGRIKKVKLQRSLEFSISSEILETI 366
VLWLLMSNLKRLQDAKLVYLVNVTIQGLELALISCCGRIKKVKLQRSLEFSISSEILETI
Sbjct: 667 VLWLLMSNLKRLQDAKLVYLVNVTIQGLELALISCCGRIKKVKLQRSLEFSISSEILETI 488
Query: 367 HERGCKVRW 375
HERGCKVRW
Sbjct: 487 HERGCKVRW 461
>TC79219 similar to GP|15810000|gb|AAL06927.1 AT5g23340/MKD15_20
{Arabidopsis thaliana}, partial (78%)
Length = 1619
Score = 97.4 bits (241), Expect = 8e-21
Identities = 61/201 (30%), Positives = 102/201 (50%), Gaps = 5/201 (2%)
Frame = +2
Query: 14 GLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVR-VSDFILQIIGSN 72
G+ ++ G L ++ +C L+ K L ++ + G R V+D IL+ + N
Sbjct: 458 GMKAIGDGLSLLHSLDVSYCRKLTDKGLSAVAKGCCDLRILHLTGCRFVTDSILEALSKN 637
Query: 73 CKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCRFVTDAAISTIANSCPN-LACL 130
C++L EL L C +T+ G+M + GC + LD+ C V+D +S+I N+C + L L
Sbjct: 638 CRNLEELVLQGCTSITDNGLMSLASGCQRIKFLDINKCSTVSDVGVSSICNACSSSLKTL 817
Query: 131 KLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRC--SKLVRLKLGLCTN 188
KL C + + + + C LE L + C V++ A+K L+ +KL L++ C N
Sbjct: 818 KLLDCYRIGDKSILSLAKFCDNLETLIIGGCRDVSNDAIKLLATACRNKLKNLRMDWCLN 997
Query: 189 ISDIGLAHIACNCPKLTELDL 209
+SD L+ I C L LD+
Sbjct: 998 VSDSSLSCILSQCRNLEALDI 1060
Score = 89.0 bits (219), Expect = 3e-18
Identities = 55/159 (34%), Positives = 81/159 (50%), Gaps = 3/159 (1%)
Frame = +2
Query: 52 SVIRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCR 110
S I I + D ++ IG L L +S C +T+ G+ V GCC+L L LT CR
Sbjct: 419 SSICITVKELPDVGMKAIGDGLSLLHSLDVSYCRKLTDKGLSAVAKGCCDLRILHLTGCR 598
Query: 111 FVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALK 170
FVTD+ + ++ +C NL L L+ C +T+ GL + S C ++ LD+ CS V+D+ +
Sbjct: 599 FVTDSILEALSKNCRNLEELVLQGCTSITDNGLMSLASGCQRIKFLDINKCSTVSDVGVS 778
Query: 171 YLSRC--SKLVRLKLGLCTNISDIGLAHIACNCPKLTEL 207
+ S L LKL C I D + +A C L L
Sbjct: 779 SICNACSSSLKTLKLLDCYRIGDKSILSLAKFCDNLETL 895
Score = 68.2 bits (165), Expect = 5e-12
Identities = 46/174 (26%), Positives = 87/174 (49%), Gaps = 6/174 (3%)
Frame = +2
Query: 2 IDVSRCNCVSPSGLLSVISG-HEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVR 60
+D+++C+ VS G+ S+ + L+ + C + + K +L + I G R
Sbjct: 734 LDINKCSTVSDVGVSSICNACSSSLKTLKLLDCYRIGDKSILSLAKFCDNLETLIIGGCR 913
Query: 61 -VSDFILQIIGSNCKS-LVELGLSKCIGVTNMGIMQVVGCC-NLTTLDLTCCRFVTDAAI 117
VS+ ++++ + C++ L L + C+ V++ + ++ C NL LD+ CC VTD A
Sbjct: 914 DVSNDAIKLLATACRNKLKNLRMDWCLNVSDSSLSCILSQCRNLEALDIGCCEEVTDTAF 1093
Query: 118 STIANSCPNLA--CLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIAL 169
I+N P L+ LK+ +C +T +G+ + C L+ LD+ C + L
Sbjct: 1094HHISNEEPGLSLKILKVSNCPKITVVGIGILLGKCSYLKYLDVRSCPHITKAGL 1255
>TC81462 similar to GP|21536497|gb|AAM60829.1 F-box protein family AtFBL4
{Arabidopsis thaliana}, partial (67%)
Length = 1452
Score = 87.4 bits (215), Expect = 8e-18
Identities = 52/166 (31%), Positives = 82/166 (49%), Gaps = 2/166 (1%)
Frame = +3
Query: 46 KNLKHLSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTL 104
K HL +++ + ++D L+ +G +C SL L L T+ G+ + GC L L
Sbjct: 879 KGCPHLKSLKLQCINLTDDALKAVGVSCLSLELLALYSFQRFTDKGLRAIGNGCKKLKNL 1058
Query: 105 DLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGV 164
L+ C F++D + IA C L L++ C + +GL +G SCL L EL L C +
Sbjct: 1059 TLSDCYFLSDKGLEAIATGCKELTHLEVNGCHNIGTLGLDSVGKSCLHLSELALLYCQRI 1238
Query: 165 NDIALKYLSR-CSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
D+ L + + C L L L C++I D + IA C L +L +
Sbjct: 1239 GDLGLLQVGKGCQFLQALHLVDCSSIGDEAMCGIATGCRNLKKLHI 1376
Score = 74.3 bits (181), Expect = 7e-14
Identities = 51/138 (36%), Positives = 72/138 (51%), Gaps = 2/138 (1%)
Frame = +3
Query: 74 KSLVELGLSKCIGVTNMGIMQVVGCC-NLTTLDLTCCRFVTDAAISTIANSCPNLACLKL 132
KSL LG++ C +T++ + V C +L TL L FV + + +A CP+L LKL
Sbjct: 735 KSLKSLGVAACAKITDISMEAVASHCGSLETLSLDS-EFVHNQGVLAVAKGCPHLKSLKL 911
Query: 133 ESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSR-CSKLVRLKLGLCTNISD 191
+ C +T+ L +G SCL LE L L D L+ + C KL L L C +SD
Sbjct: 912 Q-CINLTDDALKAVGVSCLSLELLALYSFQRFTDKGLRAIGNGCKKLKNLTLSDCYFLSD 1088
Query: 192 IGLAHIACNCPKLTELDL 209
GL IA C +LT L++
Sbjct: 1089KGLEAIATGCKELTHLEV 1142
Score = 49.3 bits (116), Expect = 2e-06
Identities = 32/97 (32%), Positives = 47/97 (47%), Gaps = 1/97 (1%)
Frame = +2
Query: 112 VTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKY 171
++D + +A+ P L LKL C VT GL + S C L+ LDL C V D L
Sbjct: 461 LSDNGLIALADGFPKLEKLKLIWCSNVTSFGLSSLASKCASLKSLDLQGCY-VGDQGLAA 637
Query: 172 L-SRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTEL 207
+ RC +L L L C ++D G I+ K+ ++
Sbjct: 638 VGQRCKQLEDLNLRFCEGLTDTGXG*ISFRRGKIIKI 748
Score = 49.3 bits (116), Expect = 2e-06
Identities = 39/125 (31%), Positives = 58/125 (46%), Gaps = 4/125 (3%)
Frame = +3
Query: 14 GLLSVISGHEGLEQINAGHC--LSELSAP-LTNGLKNLKHLSVIRIDGVRVSDFILQIIG 70
GL ++ +G + L+ + C LS+ + G K L HL V + L +G
Sbjct: 1014 GLRAIGNGCKKLKNLTLSDCYFLSDKGLEAIATGCKELTHLEVNGCHNIGTLG--LDSVG 1187
Query: 71 SNCKSLVELGLSKCIGVTNMGIMQV-VGCCNLTTLDLTCCRFVTDAAISTIANSCPNLAC 129
+C L EL L C + ++G++QV GC L L L C + D A+ IA C NL
Sbjct: 1188 KSCLHLSELALLYCQRIGDLGLLQVGKGCQFLQALHLVDCSSIGDEAMCGIATGCRNLKK 1367
Query: 130 LKLES 134
L + S
Sbjct: 1368 LHIPS 1382
Score = 33.9 bits (76), Expect = 0.10
Identities = 15/33 (45%), Positives = 21/33 (63%)
Frame = +2
Query: 177 KLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
KL +LKL C+N++ GL+ +A C L LDL
Sbjct: 503 KLEKLKLIWCSNVTSFGLSSLASKCASLKSLDL 601
>TC86421 similar to GP|2407790|gb|AAB70660.1| grr1 {Glycine max}, partial
(74%)
Length = 2856
Score = 83.6 bits (205), Expect = 1e-16
Identities = 52/146 (35%), Positives = 75/146 (50%), Gaps = 2/146 (1%)
Frame = +1
Query: 66 LQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCRFVTDAAISTIANSC 124
L+ + S C SL L V + G++++ GC L LDL C ++D A+ T+A C
Sbjct: 760 LKAVASGCPSLKSFSLWNVSSVGDEGLIEIANGCQKLEKLDLCKCPAISDKALITVAKKC 939
Query: 125 PNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVRLKLG 184
PNL L LESC + GL IG C L+ + + DC+GV D + L + LV K+
Sbjct: 940 PNLTELSLESCPSIRNEGLQAIGKFCPNLKAISIKDCAGVGDQGIAGLFSSTSLVLTKVK 1119
Query: 185 L-CTNISDIGLAHIACNCPKLTELDL 209
L +SD+ LA I +T+L L
Sbjct: 1120LQALAVSDLSLAVIGHYGKTVTDLVL 1197
Score = 69.3 bits (168), Expect = 2e-12
Identities = 61/246 (24%), Positives = 99/246 (39%), Gaps = 45/246 (18%)
Frame = +1
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLS---ELSAPLTNGLKNLKHLSV----- 53
+D+ +C +S L++V L +++ C S E + NLK +S+
Sbjct: 877 LDLCKCPAISDKALITVAKKCPNLTELSLESCPSIRNEGLQAIGKFCPNLKAISIKDCAG 1056
Query: 54 -------------------IRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQ 94
+++ + VSD L +IG K++ +L L+ V+ G
Sbjct: 1057 VGDQGIAGLFSSTSLVLTKVKLQALAVSDLSLAVIGHYGKTVTDLVLNFLPNVSERGFW- 1233
Query: 95 VVGCCN----LTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSC 150
V+G N L +L + CR VTD I + CPNL + L C +++ GL +
Sbjct: 1234 VMGNANGLHKLKSLTIASCRGVTDVGIEAVGKGCPNLKSVHLHKCAFLSDNGLISFTKAA 1413
Query: 151 LMLEELDLTDCSGVNDIA-LKYLSRC-SKLVRLKLGLCTNISDIGL------------AH 196
+ LE L L +C + L C +KL L + C I D+ L +
Sbjct: 1414 ISLESLQLEECHRITQFGFFGVLFNCGAKLKALSMISCFGIKDLDLELSPVSPCESLRSL 1593
Query: 197 IACNCP 202
CNCP
Sbjct: 1594 SICNCP 1611
Score = 60.8 bits (146), Expect = 8e-10
Identities = 34/101 (33%), Positives = 50/101 (48%), Gaps = 1/101 (0%)
Frame = +1
Query: 110 RFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIAL 169
R VT + +A+ CP+L L + V + GL +I + C LE+LDL C ++D AL
Sbjct: 739 RGVTTLGLKAVASGCPSLKSFSLWNVSSVGDEGLIEIANGCQKLEKLDLCKCPAISDKAL 918
Query: 170 -KYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
+C L L L C +I + GL I CP L + +
Sbjct: 919 ITVAKKCPNLTELSLESCPSIRNEGLQAIGKFCPNLKAISI 1041
Score = 41.6 bits (96), Expect = 5e-04
Identities = 41/174 (23%), Positives = 80/174 (45%), Gaps = 3/174 (1%)
Frame = +1
Query: 2 IDVSRCNCVSPSGLLSVISGHE-GLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVR 60
++++ V+ +GLL ++ E GL ++N C++ L+ + + L NL
Sbjct: 1669 VELTGLKGVTDAGLLPLLESSEAGLVKVNLSGCVN-LTDKVVSSLVNLH----------- 1812
Query: 61 VSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCN-LTTLDLTCCRFVTDAAIST 119
+ L+I L L CI ++N + + C L LD + C ++D+ I+
Sbjct: 1813 --GWTLEI----------LNLEGCINISNASLAAIAEHCQLLCDLDFSMCT-ISDSGITA 1953
Query: 120 IANSCP-NLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYL 172
+A++ NL L L C +VT+ L + L L++ C+ ++ A++ L
Sbjct: 1954 LAHAKQINLQILSLSGCTLVTDRSLPALRKLGHTLLGLNIQHCNSISSSAVEML 2115
>TC78335 similar to GP|16604366|gb|AAL24189.1 At1g21410/F24J8_17
{Arabidopsis thaliana}, partial (90%)
Length = 1520
Score = 76.6 bits (187), Expect = 1e-14
Identities = 58/223 (26%), Positives = 100/223 (44%), Gaps = 25/223 (11%)
Frame = +3
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGV-- 59
+D+S+ ++ L ++ G L ++N C + L + L V+ + G
Sbjct: 531 LDLSKSFKLTDRSLYAIAHGCRDLTKLNISGCSAFSDNALAYLAGFCRKLKVLNLCGCVR 710
Query: 60 RVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCRFVTDAAIS 118
SD LQ IG C L L L C V ++G+M + GC +L T+DL C ++TD ++
Sbjct: 711 AASDTALQAIGHYCNQLQSLNLGWCDKVGDVGVMSLAYGCPDLRTVDLCGCVYITDDSVI 890
Query: 119 TIANSCPNLACLKLESCDMVTEIGLYQIGSSCLM---------------LEELDLTDCSG 163
+AN CP+L L L C +T+ +Y + S + L L+++ C+
Sbjct: 891 ALANGCPHLRSLGLYFCKNITDNAMYSLAQSKVKNRMWGSVKGGNDEDGLRTLNISQCTS 1070
Query: 164 VNDIALK-------YLSRCSKLVRLKLGLCTNISDIGLAHIAC 199
+ A++ L CS L + C N++++ H AC
Sbjct: 1071LTPSAVQAVCDSSPALHTCSGRHSLIMSGCLNLTEV---HCAC 1190
Score = 61.2 bits (147), Expect = 6e-10
Identities = 48/164 (29%), Positives = 70/164 (42%), Gaps = 30/164 (18%)
Frame = +3
Query: 76 LVELGLSKCI-GVTNMGIMQVVGCCNLTTLDLTCCR-FVTDAAISTIANSCPNLACLKLE 133
L L LS C + N+ + V L TL L + + D + TIAN C +L L L
Sbjct: 363 LARLSLSWCNKNMNNLVLSLVPKFAKLQTLILRQDKPQLDDNVVGTIANFCHDLQILDLS 542
Query: 134 SCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLS-------------------- 173
+T+ LY I C L +L+++ CS +D AL YL+
Sbjct: 543 KSFKLTDRSLYAIAHGCRDLTKLNISGCSAFSDNALAYLAGFCRKLKVLNLCGCVRAASD 722
Query: 174 --------RCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
C++L L LG C + D+G+ +A CP L +DL
Sbjct: 723 TALQAIGHYCNQLQSLNLGWCDKVGDVGVMSLAYGCPDLRTVDL 854
>TC86288 similar to GP|2407790|gb|AAB70660.1| grr1 {Glycine max}, partial
(84%)
Length = 2900
Score = 70.5 bits (171), Expect = 1e-12
Identities = 57/229 (24%), Positives = 89/229 (37%), Gaps = 56/229 (24%)
Frame = +1
Query: 61 VSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCRFVTDAAIST 119
++D L+ + C SL L +++ G++++ GC + LDL ++D A+
Sbjct: 709 LTDVGLKAVAHGCPSLKSFTLWDVATISDAGLIEIANGCHQIENLDLCKLPTISDKALIA 888
Query: 120 IANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVND-----------IA 168
+A CPNL L +ESC + GL+ IG C L + + +C GV D I
Sbjct: 889 VAKHCPNLTELSIESCPSIGNEGLHAIGKLCPNLRSVSIKNCPGVRDQGIAGLLCSASII 1068
Query: 169 LKYLS--------------------------------------------RCSKLVRLKLG 184
LK L+ +L L +G
Sbjct: 1069LKKLTLESLAVSDYSLAVIGQYGFVVTDLVLNFLPNVTEKGFWVMGNGHALQQLTSLTIG 1248
Query: 185 LCTNISDIGLAHIACNCPKLTELDLYRR*WASSTDNRMQQVGHAQPSIL 233
LC ++DIGL + CP + L R + S DN + A PSI+
Sbjct: 1249LCPGVTDIGLHAVGKGCPNVKNFQLRRCSFLS--DNGLVSFTKAAPSIV 1389
Score = 61.2 bits (147), Expect = 6e-10
Identities = 52/216 (24%), Positives = 96/216 (44%), Gaps = 7/216 (3%)
Frame = +1
Query: 1 AIDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGV- 59
++ + C V+ GL +V G ++ C L + K + ++++
Sbjct: 1234 SLTIGLCPGVTDIGLHAVGKGCPNVKNFQLRRCSFLSDNGLVSFTKAAPSIVSLQLEECH 1413
Query: 60 RVSDF-ILQIIGSNCKSLVELGLSKCIGVT--NMGIMQVVGCCNLTTLDLTCCRFVTDAA 116
R++ F + I + L L L C G+ N+ + V C +++L + C V +
Sbjct: 1414 RITQFGVAGAILNRGTKLKVLTLVSCYGIKDLNLNLPAVPPCQKISSLSIRNCPGVGNFT 1593
Query: 117 ISTIANSCPNLACLKLESCDMVTEIGLYQ-IGSSCLMLEELDLTDCSGVNDIALKYLSR- 174
++ + CP L CL+L + +TE G + S L ++L+ C + D+ + + +
Sbjct: 1594 LNVLGKLCPTLQCLELIGLEGITEPGFISLLQRSKASLGNVNLSGCINLTDVGVLSMVKL 1773
Query: 175 -CSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
CS L L L C + D L IA NC L++LD+
Sbjct: 1774 HCSTLGVLNLNGCKKVGDASLTAIADNCIVLSDLDV 1881
Score = 58.5 bits (140), Expect = 4e-09
Identities = 46/166 (27%), Positives = 76/166 (45%), Gaps = 7/166 (4%)
Frame = +1
Query: 51 LSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVG----CCNLTTLDL 106
L + ++ + VSD+ L +IG + +L L+ VT G V+G LT+L +
Sbjct: 1069 LKKLTLESLAVSDYSLAVIGQYGFVVTDLVLNFLPNVTEKGFW-VMGNGHALQQLTSLTI 1245
Query: 107 TCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVND 166
C VTD + + CPN+ +L C +++ GL + + L L +C +
Sbjct: 1246 GLCPGVTDIGLHAVGKGCPNVKNFQLRRCSFLSDNGLVSFTKAAPSIVSLQLEECHRITQ 1425
Query: 167 --IALKYLSRCSKLVRLKLGLCTNISDIGLAHIAC-NCPKLTELDL 209
+A L+R +KL L L C I D+ L A C K++ L +
Sbjct: 1426 FGVAGAILNRGTKLKVLTLVSCYGIKDLNLNLPAVPPCQKISSLSI 1563
Score = 56.6 bits (135), Expect = 2e-08
Identities = 30/101 (29%), Positives = 51/101 (49%), Gaps = 1/101 (0%)
Frame = +1
Query: 110 RFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIAL 169
R +TD + +A+ CP+L L +++ GL +I + C +E LDL ++D AL
Sbjct: 703 RALTDVGLKAVAHGCPSLKSFTLWDVATISDAGLIEIANGCHQIENLDLCKLPTISDKAL 882
Query: 170 KYLSR-CSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
+++ C L L + C +I + GL I CP L + +
Sbjct: 883 IAVAKHCPNLTELSIESCPSIGNEGLHAIGKLCPNLRSVSI 1005
Score = 41.2 bits (95), Expect = 7e-04
Identities = 32/135 (23%), Positives = 66/135 (48%), Gaps = 5/135 (3%)
Frame = +1
Query: 10 VSPSGLLSVISGHEG-LEQINAGHCLSELSAPLTNGLK-NLKHLSVIRIDGVR-VSDFIL 66
++ G +S++ + L +N C++ + + +K + L V+ ++G + V D L
Sbjct: 1657 ITEPGFISLLQRSKASLGNVNLSGCINLTDVGVLSMVKLHCSTLGVLNLNGCKKVGDASL 1836
Query: 67 QIIGSNCKSLVELGLSKCIGVTNMGIMQVVG--CCNLTTLDLTCCRFVTDAAISTIANSC 124
I NC L +L +S+C +T+ GI + NL L L C V++ ++S +
Sbjct: 1837 TAIADNCIVLSDLDVSEC-AITDAGISALTRGVLFNLDVLSLAGCSLVSNKSLSALKKLG 2013
Query: 125 PNLACLKLESCDMVT 139
+L L +++C ++
Sbjct: 2014 DSLEGLNIKNCKSIS 2058
>AW684440 weakly similar to PIR|A84649|A84 probable glucose regulated
repressor protein [imported] - Arabidopsis thaliana,
partial (17%)
Length = 527
Score = 62.8 bits (151), Expect = 2e-10
Identities = 43/161 (26%), Positives = 75/161 (45%), Gaps = 4/161 (2%)
Frame = +2
Query: 53 VIRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQV-VGCCNLTTLDLTCCRF 111
++RID +R + + L L + C GVT++G+ V GC N+ L C F
Sbjct: 14 IVRIDRLRALS*FHAL-----QQLTSLTIGLCPGVTDIGLHAVGKGCPNVKNFQLRRCSF 178
Query: 112 VTDAAISTIANSCPNLACLKLESCDMVTEIGLY-QIGSSCLMLEELDLTDCSGVNDIALK 170
++D + + + P++ L+LE C +T+ G+ I + L+ L L C G+ D+ L
Sbjct: 179 LSDNGLVSFTKAAPSIVSLQLEECHRITQFGVAGAILNRGTKLKVLTLVSCYGIKDLNLN 358
Query: 171 Y--LSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
+ C + L + C + + L + CP L L+L
Sbjct: 359 LPAVPPCQTISSLSIRNCPGVGNFTLNVLGKLCPTLXCLEL 481
Score = 35.4 bits (80), Expect = 0.036
Identities = 22/66 (33%), Positives = 31/66 (46%)
Frame = +2
Query: 168 ALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDLYRR*WASSTDNRMQQVGH 227
AL +L L +GLC ++DIGL + CP + L R + S DN +
Sbjct: 38 ALS*FHALQQLTSLTIGLCPGVTDIGLHAVGKGCPNVKNFQLRRCSFLS--DNGLVSFTK 211
Query: 228 AQPSIL 233
A PSI+
Sbjct: 212 AAPSIV 229
Score = 35.0 bits (79), Expect = 0.047
Identities = 30/143 (20%), Positives = 61/143 (41%), Gaps = 4/143 (2%)
Frame = +2
Query: 1 AIDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGV- 59
++ + C V+ GL +V G ++ C L + K + ++++
Sbjct: 74 SLTIGLCPGVTDIGLHAVGKGCPNVKNFQLRRCSFLSDNGLVSFTKAAPSIVSLQLEECH 253
Query: 60 RVSDF-ILQIIGSNCKSLVELGLSKCIGVT--NMGIMQVVGCCNLTTLDLTCCRFVTDAA 116
R++ F + I + L L L C G+ N+ + V C +++L + C V +
Sbjct: 254 RITQFGVAGAILNRGTKLKVLTLVSCYGIKDLNLNLPAVPPCQTISSLSIRNCPGVGNFT 433
Query: 117 ISTIANSCPNLACLKLESCDMVT 139
++ + CP L CL+L + +T
Sbjct: 434 LNVLGKLCPTLXCLELIGLEGIT 502
>BI311086
Length = 718
Score = 52.8 bits (125), Expect = 2e-07
Identities = 50/149 (33%), Positives = 74/149 (49%), Gaps = 6/149 (4%)
Frame = +1
Query: 67 QIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTIANSCP- 125
+++ KSL L + C+ N+ IM NL L L+C +T+ I + SC
Sbjct: 79 RLVNPQFKSLF-LASAACLEDQNI-IMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRK 252
Query: 126 ----NLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLS-RCSKLVR 180
NL CL L+S + T L LE L+LT+ V+D AL +S RC L++
Sbjct: 253 IRHLNLTCLSLKS--LGTNFDLPD-------LEVLNLTNTE-VDDEALYIISNRCPALLQ 402
Query: 181 LKLGLCTNISDIGLAHIACNCPKLTELDL 209
L L C I+D G+ H+ NC +L E++L
Sbjct: 403 LVLLRCDYITDKGVMHVVNNCTQLREINL 489
Score = 46.2 bits (108), Expect = 2e-05
Identities = 32/112 (28%), Positives = 49/112 (43%), Gaps = 22/112 (19%)
Frame = +1
Query: 75 SLVELGLSKCIGVTNMGIMQVVGCCN-LTTLDLTCCRF---------------------V 112
+L +L LS +T GI ++ C + L+LTC V
Sbjct: 172 NLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEV 351
Query: 113 TDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGV 164
D A+ I+N CP L L L CD +T+ G+ + ++C L E++L C V
Sbjct: 352 DDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNV 507
>TC85594 similar to PIR|D96512|D96512 hypothetical protein F2G19.16
[imported] - Arabidopsis thaliana, partial (38%)
Length = 1334
Score = 52.8 bits (125), Expect = 2e-07
Identities = 54/206 (26%), Positives = 96/206 (46%), Gaps = 7/206 (3%)
Frame = +1
Query: 34 LSELSAPLTNGLKNLKHLSVIRIDGVRVSDFILQIIGSNCKSLVELGLS--KCIGVTNMG 91
+S++ + NL+ L +++ +D L I CK L +L + K + + G
Sbjct: 97 ISDIGLQAISNSSNLEILHLVKTP--ECTDMGLVAIAERCKLLRKLHIDGWKANRIGDEG 270
Query: 92 IMQVVGCC-NLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSC 150
++ V C NL L L T ++ +A++CPNL L L + D V + + I + C
Sbjct: 271 LIAVAKFCPNLQELVLIGVN-PTRVSLEMLASNCPNLERLALCASDTVGDPEISCIAAKC 447
Query: 151 LMLEELDLTDCSGVNDIALKYLSR-CSKLVRLKLGLCTNISDIG---LAHIACNCPKLTE 206
L L++L + C V+D+ ++ L+ C LV++K+ C ++ G L H + +
Sbjct: 448 LALKKLCIKSCP-VSDLGMEALANGCPNLVKVKVKKCKGVTPEGGDWLRHTRVSVAVNLD 624
Query: 207 LDLYRR*WASSTDNRMQQVGHAQPSI 232
D AS++D Q G PS+
Sbjct: 625 ADEAELQDASASDGGAQDNGIEFPSM 702
>TC86245 weakly similar to GP|9279671|dbj|BAB01228.1 transport inhibitor
response-like protein {Arabidopsis thaliana}, partial
(80%)
Length = 2260
Score = 52.4 bits (124), Expect = 3e-07
Identities = 47/182 (25%), Positives = 77/182 (41%), Gaps = 19/182 (10%)
Frame = +3
Query: 47 NLKHLSVIRIDGVRVSDFILQIIGSNCKSLVELGL--------SKCIGVTNMGIMQVVGC 98
NL+ L V+ V D L+ +GS C L EL + GVT G + V
Sbjct: 1434 NLRRLWVLDT----VEDKGLEAVGSYCPLLEELRVFPGDPFEEGAAHGVTESGFIAVSEG 1601
Query: 99 CNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIG----------- 147
C L CR +T+AA++T+ +CP+ +L C M YQ G
Sbjct: 1602 CRKLHYVLYFCRQMTNAAVATVVENCPDFTHFRL--CIMTPGQPDYQTGEPMDEAFGAVV 1775
Query: 148 SSCLMLEELDLTDCSGVNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTEL 207
+C L+ L ++ + D+ +Y+ + +K + SD + + CPKL +L
Sbjct: 1776 KTCTKLQRLAVS--GSLTDLTFEYIGKYAKNLETLSVAFAGSSDWAMQCVLVGCPKLRKL 1949
Query: 208 DL 209
++
Sbjct: 1950 EI 1955
Score = 40.4 bits (93), Expect = 0.001
Identities = 23/62 (37%), Positives = 37/62 (59%)
Frame = +3
Query: 112 VTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKY 171
V+D ++ +A S PN L L SCD + GL + ++C L ELD+ + +GV+D + +
Sbjct: 840 VSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNCKNLTELDIQE-NGVDDKSGNW 1016
Query: 172 LS 173
LS
Sbjct: 1017LS 1022
Score = 33.9 bits (76), Expect = 0.10
Identities = 24/58 (41%), Positives = 32/58 (54%), Gaps = 1/58 (1%)
Frame = +3
Query: 153 LEELDLTDCSGVNDIALKYLS-RCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
LEEL L + V+D +L++L+ L L C S GLA +A NC LTELD+
Sbjct: 810 LEELRLKRMA-VSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNCKNLTELDI 980
>BE319841 weakly similar to GP|21592320|gb| unknown {Arabidopsis thaliana},
partial (10%)
Length = 363
Score = 51.6 bits (122), Expect = 5e-07
Identities = 38/113 (33%), Positives = 55/113 (48%), Gaps = 2/113 (1%)
Frame = +1
Query: 79 LGLSKCIGVTNMGIMQVVG-CCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDM 137
L LS C + GI+QV+ CCN+ L+L+ C V I P L L L S
Sbjct: 13 LDLSNCCRIFEEGIVQVLRMCCNIRHLNLSKCSIVR----LEIDFEVPKLEVLNL-SYTK 177
Query: 138 VTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKY-LSRCSKLVRLKLGLCTNI 189
V + LY I SC L +L L DC+ V +K+ + C++L ++ L C +
Sbjct: 178 VDDEALYMISKSCCGLLKLSLQDCNDVTKKGVKHVVENCTQLRKISLNGCFKV 336
>TC80249 weakly similar to GP|10716947|gb|AAG21976.1 SKP1 interacting
partner 1 {Arabidopsis thaliana}, partial (72%)
Length = 1197
Score = 50.1 bits (118), Expect = 1e-06
Identities = 26/77 (33%), Positives = 42/77 (53%), Gaps = 1/77 (1%)
Frame = +1
Query: 110 RFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIAL 169
R +D +++ +A CPNL L + SC VT+ + +I + C L ELD++ C + +L
Sbjct: 328 RHCSDRSLALVAQRCPNLEVLSIRSCPRVTDDSMSKIATGCPNLRELDISYCYEITHESL 507
Query: 170 KYLSR-CSKLVRLKLGL 185
+ R CS + LK L
Sbjct: 508 VLIGRNCSNIKVLKRNL 558
Score = 45.8 bits (107), Expect = 3e-05
Identities = 41/133 (30%), Positives = 55/133 (40%), Gaps = 21/133 (15%)
Frame = +1
Query: 98 CCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELD 157
C NL L + C VTD ++S IA CPNL L + C +T L IG +C ++ L
Sbjct: 370 CPNLEVLSIRSCPRVTDDSMSKIATGCPNLRELDISYCYEITHESLVLIGRNCSNIKVLK 549
Query: 158 ------LTDCSGVNDIALKYLSRC------------SKLVRLKLGLCTNISDI---GLAH 196
L V + YL+ C + + L+ GL S + GL
Sbjct: 550 RNLMNWLDPSQHVGIVPDDYLNACPQDGDSEAAAIANSMPHLE-GLEIRFSKLTAKGLNS 726
Query: 197 IACNCPKLTELDL 209
I CP L LDL
Sbjct: 727 ICQGCPNLEFLDL 765
Score = 45.8 bits (107), Expect = 3e-05
Identities = 42/183 (22%), Positives = 73/183 (38%), Gaps = 25/183 (13%)
Frame = +1
Query: 50 HLSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTLDLTC 108
H+ + +I SD L ++ C +L L + C VT+ + ++ GC NL LD++
Sbjct: 301 HIFLTQIRIRHCSDRSLALVAQRCPNLEVLSIRSCPRVTDDSMSKIATGCPNLRELDISY 480
Query: 109 CRFVTDAAISTIANSCPNLACLK--------------------LESCDMVTEIGLYQIGS 148
C +T ++ I +C N+ LK L +C + I +
Sbjct: 481 CYEITHESLVLIGRNCSNIKVLKRNLMNWLDPSQHVGIVPDDYLNACPQDGDSEAAAIAN 660
Query: 149 SCLMLEELDL----TDCSGVNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKL 204
S LE L++ G+N I C L L L C N++ +A + + L
Sbjct: 661 SMPHLEGLEIRFSKLTAKGLNSIC----QGCPNLEFLDLSGCANLTSRDIAKASSSLSHL 828
Query: 205 TEL 207
++
Sbjct: 829 KDI 837
Score = 34.3 bits (77), Expect = 0.080
Identities = 19/59 (32%), Positives = 30/59 (50%)
Frame = +1
Query: 151 LMLEELDLTDCSGVNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
+ L ++ + CS +AL RC L L + C ++D ++ IA CP L ELD+
Sbjct: 304 IFLTQIRIRHCSD-RSLAL-VAQRCPNLEVLSIRSCPRVTDDSMSKIATGCPNLRELDI 474
Score = 27.7 bits (60), Expect = 7.5
Identities = 32/154 (20%), Positives = 62/154 (39%), Gaps = 24/154 (15%)
Frame = +1
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHC-----------------LSELSAPLTNG 44
+ + C V+ + + +G L +++ +C + L L N
Sbjct: 388 LSIRSCPRVTDDSMSKIATGCPNLRELDISYCYEITHESLVLIGRNCSNIKVLKRNLMNW 567
Query: 45 LKNLKHLSVIRIDGVRV----SDFILQIIGSNCKSL--VELGLSKCIGVTNMGIMQVV-G 97
L +H+ ++ D + D I ++ L +E+ SK +T G+ + G
Sbjct: 568 LDPSQHVGIVPDDYLNACPQDGDSEAAAIANSMPHLEGLEIRFSK---LTAKGLNSICQG 738
Query: 98 CCNLTTLDLTCCRFVTDAAISTIANSCPNLACLK 131
C NL LDL+ C +T I+ ++S +L +K
Sbjct: 739 CPNLEFLDLSGCANLTSRDIAKASSSLSHLKDIK 840
>TC86171 weakly similar to GP|13507547|gb|AAK28636.1 unknown protein
{Arabidopsis thaliana}, partial (89%)
Length = 2343
Score = 49.3 bits (116), Expect = 2e-06
Identities = 56/200 (28%), Positives = 93/200 (46%), Gaps = 7/200 (3%)
Frame = +1
Query: 1 AIDVSRCNCVSPSGLLSVISGHEGLEQINAGHC--LSELSAPLTNGLKNLKHLSV----- 53
A+DVS N V+ GL ++ L+ + +C SE +GL NL LS+
Sbjct: 631 AVDVSGSN-VTDHGL-RLLKDCLNLQALTLNYCDQFSEHGLKHLSGLSNLTSLSIRKSCA 804
Query: 54 IRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVT 113
+ DG+R SN +L +L L +C + G + G L +L++ CC+ VT
Sbjct: 805 VTPDGMRAF--------SNLVNLEKLDLERCSDIHG-GFVHFKGLKKLESLNIGCCKCVT 957
Query: 114 DAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLS 173
D+ + I+ NL L++ S +T++G+ + L L++ CS + +Y+S
Sbjct: 958 DSDMKAISGFI-NLKELQI-SNSSITDLGISYL-RGLQKLSTLNVEGCS-ITAACFEYIS 1125
Query: 174 RCSKLVRLKLGLCTNISDIG 193
+ L L L C +SD G
Sbjct: 1126ALAALACLNLNRC-GLSDDG 1182
>BQ751306 similar to GP|18376293|em related to protein GRR1 {Neurospora
crassa}, partial (13%)
Length = 371
Score = 47.8 bits (112), Expect = 7e-06
Identities = 35/127 (27%), Positives = 62/127 (48%), Gaps = 1/127 (0%)
Frame = -3
Query: 84 CIGVTNMGIMQVVGCCN-LTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIG 142
C +T+ + ++V CN + +DL CC +TD +++ +A P L + L C +T+
Sbjct: 366 CGHITDEAVKRLVQACNRIRYIDLGCCTLLTDDSVTRLAQ-LPKLKRIGLVKCSNITDES 190
Query: 143 LYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCP 202
++ + + D SG D +Y + S L R+ L CTN++ + + CP
Sbjct: 189 VFALARA--NHRPRARRDASGHID---EYYA--SSLERVHLSYCTNLTLKSIIKLQNYCP 31
Query: 203 KLTELDL 209
+LT L L
Sbjct: 30 RLTHLSL 10
>BI312046 similar to GP|17979057|gb unknown protein {Arabidopsis thaliana},
partial (37%)
Length = 621
Score = 46.6 bits (109), Expect = 2e-05
Identities = 38/144 (26%), Positives = 65/144 (44%), Gaps = 2/144 (1%)
Frame = +2
Query: 66 LQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCC-NLTTLDLTCCRFVT-DAAISTIANS 123
++ IG C+ L EL L C + G M V C NL TL L C+ + + + S
Sbjct: 17 IKAIGKCCQMLEELTL--CDHRMDDGWMAAVSYCENLKTLRLQSCKKIDLNPGLDEYLGS 190
Query: 124 CPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVRLKL 183
CP L L L C + + + + S C + E+ + DC G ++ + + C ++ L
Sbjct: 191 CPALERLHLLKCQLRDKKSVSALFSVCRVAREIIIQDCWGFDNGMFGFATVCRRVKLFYL 370
Query: 184 GLCTNISDIGLAHIACNCPKLTEL 207
C+ ++ GL + + +L L
Sbjct: 371 EGCSLLTTEGLESVIHSWKELQSL 442
>TC84223 weakly similar to GP|18252175|gb|AAL61920.1 unknown protein
{Arabidopsis thaliana}, partial (90%)
Length = 790
Score = 45.4 bits (106), Expect = 3e-05
Identities = 26/95 (27%), Positives = 49/95 (51%), Gaps = 1/95 (1%)
Frame = +3
Query: 65 ILQIIGSNC-KSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTIANS 123
+++I + C +L + L G+T+ G+++++ F+TD ++ IA S
Sbjct: 387 LIRISYAKCINNLTSISLWGLTGITDEGVVKLISRTKSLRHLNVGGTFITDESLFAIARS 566
Query: 124 CPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDL 158
CP + + L SC VTE GL + CL L+ +++
Sbjct: 567 CPKMETIVLWSCRHVTENGLIALVDQCLKLKSMNV 671
>TC76362 similar to GP|10716949|gb|AAG21977.1 SKP1 interacting partner 2
{Arabidopsis thaliana}, partial (57%)
Length = 1307
Score = 45.1 bits (105), Expect = 5e-05
Identities = 26/70 (37%), Positives = 38/70 (54%)
Frame = +1
Query: 98 CCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELD 157
C NLT L L CR VT+ + +A +C NL L + SC + G+ + + +LEEL
Sbjct: 505 CKNLTRLKLRACREVTEIGMLGLARNCKNLKKLSVVSC-LFGVKGIRAVVDNSNVLEELS 681
Query: 158 LTDCSGVNDI 167
+ GVND+
Sbjct: 682 VKRLRGVNDV 711
Score = 42.4 bits (98), Expect = 3e-04
Identities = 23/65 (35%), Positives = 36/65 (55%), Gaps = 3/65 (4%)
Frame = +1
Query: 100 NLTTLDLTCCR---FVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEEL 156
++T L L C + + D A+ I+ C NL LKL +C VTEIG+ + +C L++L
Sbjct: 424 SVTKLALRCNKKSTSINDDALILISLRCKNLTRLKLRACREVTEIGMLGLARNCKNLKKL 603
Query: 157 DLTDC 161
+ C
Sbjct: 604 SVVSC 618
Score = 40.4 bits (93), Expect = 0.001
Identities = 20/49 (40%), Positives = 30/49 (60%), Gaps = 1/49 (2%)
Frame = +1
Query: 162 SGVNDIALKYLS-RCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
+ +ND AL +S RC L RLKL C +++IG+ +A NC L +L +
Sbjct: 463 TSINDDALILISLRCKNLTRLKLRACREVTEIGMLGLARNCKNLKKLSV 609
>TC81941 similar to GP|15420162|gb|AAK97303.1 F-box containing protein ORE9
{Arabidopsis thaliana}, partial (35%)
Length = 1017
Score = 44.3 bits (103), Expect = 8e-05
Identities = 30/97 (30%), Positives = 51/97 (51%), Gaps = 5/97 (5%)
Frame = +2
Query: 108 CCRFVTDA--AISTIANSCPNLACLKLESCDMVT-EIGLYQIGSS-CLMLEELDLTDCSG 163
CC+ V + A+ ++ CPNL +KL + IG G + C L+ L + C
Sbjct: 2 CCKNVXEXSFALEMLSFKCPNLKVVKLGQFQGICLAIGSRLDGIALCHGLQSLSVNTCGD 181
Query: 164 VNDIALKYLSR-CSKLVRLKLGLCTNISDIGLAHIAC 199
++D+ L + R CS+LVR ++ C +++ GL +AC
Sbjct: 182 LDDMGLIEIGRGCSRLVRFEIQGCKLVTEKGLRTMAC 292
>TC81565 similar to GP|6041850|gb|AAF02159.1| unknown protein {Arabidopsis
thaliana}, partial (12%)
Length = 1070
Score = 42.7 bits (99), Expect = 2e-04
Identities = 28/88 (31%), Positives = 46/88 (51%), Gaps = 1/88 (1%)
Frame = +3
Query: 126 NLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIAL-KYLSRCSKLVRLKLG 184
N + L + D+ T + ++++ LE L L C+ ++D L + LS S L +L L
Sbjct: 717 NPSSLSAKGLDVYT-VHIHRLLRRFQHLESLSLCGCTELDDSXLTRLLSYGSNLQKLNLD 893
Query: 185 LCTNISDIGLAHIACNCPKLTELDLYRR 212
C +++ L+ A CP LT + LYRR
Sbjct: 894 CCWKVTNYWLSLTASGCPSLTTIXLYRR 977
Score = 41.6 bits (96), Expect = 5e-04
Identities = 27/92 (29%), Positives = 46/92 (49%), Gaps = 1/92 (1%)
Frame = +3
Query: 76 LVELGLSKCIGVTNMGIMQVVGC-CNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLES 134
L L L C + + + +++ NL L+L CC VT+ +S A+ CP+L + L
Sbjct: 795 LESLSLCGCTELDDSXLTRLLSYGSNLQKLNLDCCWKVTNYWLSLTASGCPSLTTIXLYR 974
Query: 135 CDMVTEIGLYQIGSSCLMLEELDLTDCSGVND 166
++T+ GL ++CL L+ CS +D
Sbjct: 975 RLIITDEGLETXTTACLSXXCXXLSYCSXXSD 1070
Score = 40.4 bits (93), Expect = 0.001
Identities = 27/98 (27%), Positives = 45/98 (45%), Gaps = 2/98 (2%)
Frame = +3
Query: 45 LKNLKHLSVIRIDG-VRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGI-MQVVGCCNLT 102
L+ +HL + + G + D L + S +L +L L C VTN + + GC +LT
Sbjct: 777 LRRFQHLESLSLCGCTELDDSXLTRLLSYGSNLQKLNLDCCWKVTNYWLSLTASGCPSLT 956
Query: 103 TLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTE 140
T+ L +TD + T +C + C L C ++
Sbjct: 957 TIXLYRRLIITDEGLETXTTACLSXXCXXLSYCSXXSD 1070
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.336 0.145 0.470
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,720,498
Number of Sequences: 36976
Number of extensions: 207222
Number of successful extensions: 1800
Number of sequences better than 10.0: 87
Number of HSP's better than 10.0 without gapping: 1688
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1755
length of query: 375
length of database: 9,014,727
effective HSP length: 98
effective length of query: 277
effective length of database: 5,391,079
effective search space: 1493328883
effective search space used: 1493328883
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144541.17


