

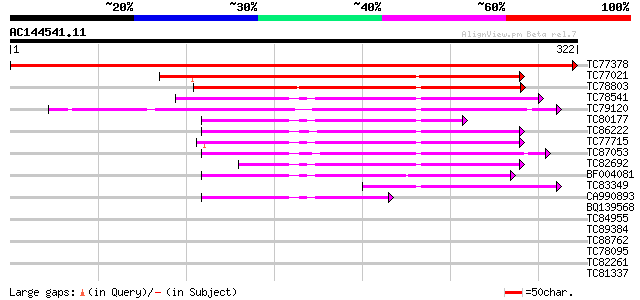
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144541.11 - phase: 0
(322 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77378 similar to PIR|D86223|D86223 hypothetical protein [impor... 635 0.0
TC77021 similar to GP|5360595|dbj|BAA82069.1 nClpP5 {Arabidopsis... 168 3e-42
TC78803 similar to GP|19698857|gb|AAL91164.1 unknown protein {Ar... 162 2e-40
TC78541 similar to GP|13899067|gb|AAK48955.1 ATP-dependent Clp p... 114 4e-26
TC79120 similar to GP|14596089|gb|AAK68772.1 ATP-dependent Clp p... 112 1e-25
TC80177 homologue to GP|9759372|dbj|BAB09831.1 ATP-dependent pro... 100 1e-21
TC86222 similar to GP|9968665|emb|CAC06126.1 putative ATP-depend... 99 2e-21
TC77715 homologue to GP|15485610|emb|CAC67407. Clp protease 2 pr... 95 4e-20
TC87053 similar to PIR|T52454|T52454 ATP-dependent Clp proteinas... 94 7e-20
TC82692 homologue to PIR|T52455|T52455 ATP-dependent clp protein... 94 7e-20
BF004081 similar to SP|P12210|CLPP ATP-dependent Clp protease pr... 79 2e-15
TC83349 similar to GP|9759372|dbj|BAB09831.1 ATP-dependent prote... 66 2e-11
CA990893 similar to SP|P30063|CLPP ATP-dependent Clp protease pr... 54 8e-08
BQ139568 similar to SP|P12210|CLPP_ ATP-dependent Clp protease p... 40 0.001
TC84955 similar to GP|9716226|emb|CAC01587.1 putative lipoprotei... 34 0.066
TC89384 similar to GP|15450417|gb|AAK96502.1 At1g26300/F28B23_4 ... 33 0.19
TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Ar... 31 0.73
TC78095 weakly similar to PIR|G84776|G84776 hypothetical protein... 31 0.73
TC82261 similar to GP|10177249|dbj|BAB10717. contains similarity... 31 0.73
TC81337 similar to GP|15723293|gb|AAL06332.1 U2 auxiliary factor... 30 1.6
>TC77378 similar to PIR|D86223|D86223 hypothetical protein [imported] -
Arabidopsis thaliana, partial (54%)
Length = 1372
Score = 635 bits (1637), Expect = 0.0
Identities = 322/322 (100%), Positives = 322/322 (100%)
Frame = +1
Query: 1 MATCFRVPISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESS 60
MATCFRVPISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESS
Sbjct: 142 MATCFRVPISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESS 321
Query: 61 PETLFNPSSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPPPDLPSLLLNGRI 120
PETLFNPSSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPPPDLPSLLLNGRI
Sbjct: 322 PETLFNPSSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPPPDLPSLLLNGRI 501
Query: 121 VYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYDA 180
VYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYDA
Sbjct: 502 VYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYDA 681
Query: 181 MMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVLI 240
MMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVLI
Sbjct: 682 MMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVLI 861
Query: 241 HAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVIDKILWRGQEKIM 300
HAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVIDKILWRGQEKIM
Sbjct: 862 HAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVIDKILWRGQEKIM 1041
Query: 301 ADVPSRDDRENGTAGAKVENRF 322
ADVPSRDDRENGTAGAKVENRF
Sbjct: 1042ADVPSRDDRENGTAGAKVENRF 1107
>TC77021 similar to GP|5360595|dbj|BAA82069.1 nClpP5 {Arabidopsis thaliana},
partial (65%)
Length = 1521
Score = 168 bits (425), Expect = 3e-42
Identities = 91/213 (42%), Positives = 134/213 (62%), Gaps = 6/213 (2%)
Frame = +1
Query: 86 MATPAQLERSASYSQQR------RPKRPPPDLPSLLLNGRIVYIGMPLVPAVTELVIAEL 139
M T A S S S R RP+ PPDLPSLLL+ RI Y+GMP+VPAVTEL++A+L
Sbjct: 397 MDTNASSRHSMSVSMYRGGRGTGRPRNAPPDLPSLLLDSRICYLGMPIVPAVTELILAQL 576
Query: 140 MFLQWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQ 199
M+L + P +P+Y+YINS+GT ETV E++ ++I D + +K+++ TV L A GQ
Sbjct: 577 MWLDYDNPAKPVYVYINSSGTQNEKNETVGSETDAYSIADMISHIKSDVYTVNLAMAYGQ 756
Query: 200 ACLLLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKH 259
A ++LS G KG R + PH+ A + P+V S +D+ I AKE+ N + ++LLAK
Sbjct: 757 AAMILSLGKKGYRAVLPHSSAKVFLPKVHRSS-GSVADMWIKAKELEANSEYYIELLAKG 933
Query: 260 TENSEETVSNVMKRSYYMDALLAKEFGVIDKIL 292
T S+E ++ ++R+ Y A E+G+ DKI+
Sbjct: 934 TGKSKEEIAKDVQRTRYFQPQDAIEYGLADKIM 1032
>TC78803 similar to GP|19698857|gb|AAL91164.1 unknown protein {Arabidopsis
thaliana}, partial (79%)
Length = 1200
Score = 162 bits (409), Expect = 2e-40
Identities = 88/189 (46%), Positives = 122/189 (63%)
Frame = +2
Query: 105 KRPPPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRAD 164
++PPPDL S L RIVY+GM LVP+VTEL++AE ++LQ+ +PIY+YINSTGTT+
Sbjct: 320 EQPPPDLASYLYKNRIVYLGMSLVPSVTELMVAEFLYLQYDDETKPIYMYINSTGTTKG- 496
Query: 165 GETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQ 224
GE + E+E A+YD M +K I T+ +G A G+A LLL+AG KG R P + MI+Q
Sbjct: 497 GEKLGYETEALAVYDLMRYIKPPIFTLCVGNAWGEAALLLAAGAKGNRSALPSSTIMIRQ 676
Query: 225 PRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKE 284
P G A+DV + KE+ + LV LLAKHTE + E + ++R Y A E
Sbjct: 677 PIARFQG--QATDVNLARKEITRVKTELVNLLAKHTEKTPEQIEADIRRPKYFSPTEAVE 850
Query: 285 FGVIDKILW 293
+G+IDK+L+
Sbjct: 851 YGIIDKVLY 877
>TC78541 similar to GP|13899067|gb|AAK48955.1 ATP-dependent Clp protease;
nClpP3 {Arabidopsis thaliana}, partial (64%)
Length = 1158
Score = 114 bits (286), Expect = 4e-26
Identities = 70/209 (33%), Positives = 115/209 (54%)
Frame = +3
Query: 95 SASYSQQRRPKRPPPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIY 154
SA + PK D ++LL RIV++G + + VI++L+FL P + I ++
Sbjct: 240 SAKTASPWLPKFEELDTTNMLLRQRIVFLGSQVDDMTADFVISQLLFLDADDPTKDIKLF 419
Query: 155 INSTGTTRADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYM 214
INS G G A G +YDAM K +++TV LG A LL+AGTKG+RY
Sbjct: 420 INSPG-----GSVTA----GMGVYDAMKLCKADVSTVCLGLAASMGAFLLAAGTKGKRYC 572
Query: 215 TPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRS 274
P+++ MI QP + G A+++ I A+E+M ++ + K+L++ T EE + R
Sbjct: 573 MPNSRVMIHQPLGSAGG--KATEMSIRARELMYHKIKINKILSRITGKPEEQIELDTDRD 746
Query: 275 YYMDALLAKEFGVIDKILWRGQEKIMADV 303
+M+ AKE+G+ID ++ G+ ++A +
Sbjct: 747 NFMNPWEAKEYGLIDDVIDDGKPGLVAPI 833
>TC79120 similar to GP|14596089|gb|AAK68772.1 ATP-dependent Clp
protease-like protein {Arabidopsis thaliana}, partial
(67%)
Length = 1110
Score = 112 bits (281), Expect = 1e-25
Identities = 83/291 (28%), Positives = 143/291 (48%)
Frame = +1
Query: 23 TPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLFNPSSSPDNLPFLDIFDS 82
+P+ R + T+ SK I + P S ++P + SP P +
Sbjct: 25 SPSLSRRIPTL--FSKPTISSFNSKPHFPLQFSQFRTTPINCVLTTPSPSKNPNFN---- 186
Query: 83 PQLMATPAQLERSASYSQQRRPKRPPPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFL 142
PQL + S+ + +R + D LLL RIV++G + V + ++++L+ L
Sbjct: 187 PQLQKPSITFQLSSPQTPERAIRGAESDTMGLLLRERIVFLGSEIDDFVADAIMSQLLLL 366
Query: 143 QWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACL 202
P + I ++INS G + S AIYDA+ ++ +++T+A+G + A +
Sbjct: 367 DAQDPTKDIKLFINSPGGSL---------SATMAIYDAVQLVRADVSTIAMGISASTASI 519
Query: 203 LLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTEN 262
+L GTKG+R P+ + MI QP +SG A DV I AKE+M N++ + ++++ T
Sbjct: 520 ILGGGTKGKRLAMPNTRIMIAQPPGGASG--QAIDVEIQAKEIMHNKNNVSRIISSFTGR 693
Query: 263 SEETVSNVMKRSYYMDALLAKEFGVIDKILWRGQEKIMADVPSRDDRENGT 313
S E V + R Y+ + A E+G+ID ++ R E I+ +P + E T
Sbjct: 694 SLEQVLKDIDRDRYLSPIEAVEYGIIDGVIDR--EMIIPLLPVPEKVEEAT 840
>TC80177 homologue to GP|9759372|dbj|BAB09831.1 ATP-dependent protease
proteolytic subunit ClpP-like protein {Arabidopsis
thaliana}, partial (66%)
Length = 648
Score = 100 bits (248), Expect = 1e-21
Identities = 62/151 (41%), Positives = 83/151 (54%)
Frame = +2
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RIV I P+ +V+A+L+FL+ P +PI +Y+NS G G A
Sbjct: 215 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPG-----GAVTA 379
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
G AIYD M +++ INT+ LG A LLL AG KG+R P+A MI QP
Sbjct: 380 ----GLAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGY 547
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHT 260
SG A D+ IH K+++ D L +L KHT
Sbjct: 548 SG--QAKDIAIHTKQIVXMWDALNELYVKHT 634
>TC86222 similar to GP|9968665|emb|CAC06126.1 putative ATP-dependent clp
serine protease proteolytic subunit {Pisum sativum},
complete
Length = 1054
Score = 99.4 bits (246), Expect = 2e-21
Identities = 64/183 (34%), Positives = 94/183 (50%)
Frame = +3
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
DL S+L RI++IG P+ V + VI++L+ L + P I +YIN G G T +
Sbjct: 363 DLSSVLFRNRIIFIGQPVNSQVAQRVISQLVTLATIDPDADILMYINCPG-----GSTYS 527
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
+ AIYD M +K ++ TV G A Q LLL+ G KG RY P+A+ MI QP+
Sbjct: 528 V----LAIYDCMSWIKPKVGTVCFGVAASQGTLLLAGGEKGMRYSMPNARIMINQPQCGF 695
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G DV E + +R + K+ + T + E V +R ++ A EFG+ID
Sbjct: 696 GG--HVEDVRRLVNESVQSRYKMEKMFSAFTGQALEKVQEYTERDSFLSVSEALEFGLID 869
Query: 290 KIL 292
+L
Sbjct: 870 GVL 878
>TC77715 homologue to GP|15485610|emb|CAC67407. Clp protease 2 proteolytic
subunit {Lycopersicon esculentum}, partial (76%)
Length = 1149
Score = 94.7 bits (234), Expect = 4e-20
Identities = 59/193 (30%), Positives = 102/193 (52%), Gaps = 7/193 (3%)
Frame = +3
Query: 107 PPP-------DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTG 159
PPP + S L RI+ G + + +++++L++L + P + I +Y+NS G
Sbjct: 372 PPPMVQERFQSVISQLFQYRIIRCGGAVDDDMANIIVSQLLYLDAVDPNKDIVMYVNSPG 551
Query: 160 TTRADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAK 219
G A G AI+D M ++ +++TV +G A LLSAGTKG+R+ P+++
Sbjct: 552 -----GSVTA----GMAIFDTMKHIRPDVSTVCVGLAASMGAFLLSAGTKGKRFSLPNSR 704
Query: 220 AMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDA 279
MI QP G +D+ I A E++ ++ L L+ T S + ++ R ++M A
Sbjct: 705 IMIHQPLGGFQG--GQTDIDIQANEMLHHKANLNGYLSYQTGQSLKKINQDTDRDFFMSA 878
Query: 280 LLAKEFGVIDKIL 292
AKE+G+ID ++
Sbjct: 879 KEAKEYGLIDGVI 917
>TC87053 similar to PIR|T52454|T52454 ATP-dependent Clp proteinase (EC
3.4.21.-) catalytic chain P2 [imported] - Arabidopsis
thaliana, partial (74%)
Length = 1135
Score = 94.0 bits (232), Expect = 7e-20
Identities = 65/198 (32%), Positives = 101/198 (50%)
Frame = +3
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
DL + L R+++IG + ++ ++A L++L + + +Y+YIN G G+
Sbjct: 372 DLWNALYRERVIFIGQEIDEELSNQILATLLYLDSVDNSKKLYLYINGPG-----GDLTP 536
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
S +YD M ++T I T +G A G A LL+AG KG R P A+ +IQ P +
Sbjct: 537 CMS----LYDTMQSLQTPICTHCIGQAYGLAAFLLAAGEKGNRTAMPLARIVIQSPAGAA 704
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G A D+ A E++ RD L LA T E ++ +KR DA A ++G+ID
Sbjct: 705 RG--RADDIQNEAAELLRIRDYLFTELANKTGQPVEKITEDLKRVKRFDAQEALDYGLID 878
Query: 290 KILWRGQEKIMADVPSRD 307
KI+ +I AD P +D
Sbjct: 879 KIV--RPRRIKADAPRKD 926
>TC82692 homologue to PIR|T52455|T52455 ATP-dependent clp proteinase (EC
3.4.21.-) chain P1 [imported] - Arabidopsis thaliana,
partial (73%)
Length = 1115
Score = 94.0 bits (232), Expect = 7e-20
Identities = 53/162 (32%), Positives = 91/162 (55%)
Frame = +2
Query: 131 VTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYDAMMQMKTEINT 190
+ +++++L++L + P + I +Y+NS G G A G AI+D M ++ +++T
Sbjct: 455 MANIIVSQLLYLDAVDPSKDIVMYVNSPG-----GSVTA----GMAIFDTMRHIRPDVST 607
Query: 191 VALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRD 250
V +G A LLSAGTKG+RY P+++ MI QP G +D+ I A E++ ++
Sbjct: 608 VCVGLAASMGAFLLSAGTKGKRYSLPNSRIMIHQPLGGFQG--GQTDIDIQANEMLHHKA 781
Query: 251 TLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVIDKIL 292
L L+ T S E ++ R ++M A AKE+G+ID ++
Sbjct: 782 NLNGYLSYQTGQSLEKINQDTDRDFFMSAKEAKEYGLIDGVI 907
>BF004081 similar to SP|P12210|CLPP ATP-dependent Clp protease proteolytic
subunit (EC 3.4.21.92) (Endopeptidase Clp). [Common
tobacco], partial (96%)
Length = 574
Score = 79.0 bits (193), Expect = 2e-15
Identities = 48/178 (26%), Positives = 90/178 (49%)
Frame = -2
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
DL + L R++++G + ++ ++ +++L +Y++INS G GE ++
Sbjct: 504 DLYNRLFQERLLFLGQEVNTEISNQLVGLMVYLSLEDKTRDLYLFINSPG-----GEVIS 340
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
G AI+D M ++ E++TV +G A LLL G +R PHA+ MI QP S
Sbjct: 339 ----GMAIFDIMQVVEAEVHTVCVGLAASMGSLLLVGGEFTKRLAFPHARVMIHQP-ATS 175
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGV 287
+ ++ A+E++ R T+ + A+ T + + M+R +M A A+ +G+
Sbjct: 174 LYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEEAEAYGI 1
>TC83349 similar to GP|9759372|dbj|BAB09831.1 ATP-dependent protease
proteolytic subunit ClpP-like protein {Arabidopsis
thaliana}, partial (43%)
Length = 814
Score = 66.2 bits (160), Expect = 2e-11
Identities = 37/113 (32%), Positives = 60/113 (52%)
Frame = +1
Query: 201 CLLLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHT 260
C AG K + P+A MI QP SG A D+ IH K+++ D L +L KHT
Sbjct: 55 CFQRMAGAKHQTRALPNATIMIHQPSGGYSG--QAKDIAIHTKQIVRMWDALNELYVKHT 228
Query: 261 ENSEETVSNVMKRSYYMDALLAKEFGVIDKILWRGQEKIMADVPSRDDRENGT 313
+ + M R Y+M A AKEFG+ID+++ + +++D + + ++ G+
Sbjct: 229 GQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLVSDAVADEGKDKGS 387
>CA990893 similar to SP|P30063|CLPP ATP-dependent Clp protease proteolytic
subunit (EC 3.4.21.92) (Endopeptidase Clp).
[Beechdrops], partial (61%)
Length = 464
Score = 53.9 bits (128), Expect = 8e-08
Identities = 31/109 (28%), Positives = 56/109 (50%)
Frame = +1
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
DL + L R++++G + ++ ++ +++L +Y++INS G GE ++
Sbjct: 133 DLYNRLFQERLLFLGQEVNTEISNQLVGLMVYLSLEDNTRDLYLFINSPG-----GEVIS 297
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHA 218
G AI+D M ++ E++TV +G A LLL G +R PHA
Sbjct: 298 ----GMAIFDIMQVVEAEVHTVCVGLAASMGSLLLVGGEFTKRLAFPHA 432
>BQ139568 similar to SP|P12210|CLPP_ ATP-dependent Clp protease proteolytic
subunit (EC 3.4.21.92) (Endopeptidase Clp). [Common
tobacco], partial (41%)
Length = 714
Score = 40.4 bits (93), Expect = 0.001
Identities = 24/89 (26%), Positives = 44/89 (48%)
Frame = -3
Query: 124 GMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYDAMMQ 183
G + ++ ++ +++L +Y++INS G GE ++ G AI+D M
Sbjct: 589 GQEVNTEISNQLVGLMVYLSLEDKTRDLYLFINSPG-----GEVIS----GMAIFDIMQV 437
Query: 184 MKTEINTVALGAAVGQACLLLSAGTKGRR 212
++ E++TV +G A LLL G +R
Sbjct: 436 VEAEVHTVCVGLAASMGSLLLVGGEFTKR 350
>TC84955 similar to GP|9716226|emb|CAC01587.1 putative lipoprotein
{Streptomyces coelicolor A3(2)}, partial (2%)
Length = 650
Score = 34.3 bits (77), Expect = 0.066
Identities = 20/64 (31%), Positives = 29/64 (45%)
Frame = -1
Query: 8 PISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLFNP 67
P + MPASSTR + T LR LR ++++ + P P F ++ V L P
Sbjct: 479 PSTNSAPMPASSTRTSSTALRPLRRLSSLRSGRTPRTPGTGFPARSVLVLMLPRRRLARP 300
Query: 68 SSSP 71
P
Sbjct: 299 RRRP 288
>TC89384 similar to GP|15450417|gb|AAK96502.1 At1g26300/F28B23_4
{Arabidopsis thaliana}, partial (46%)
Length = 801
Score = 32.7 bits (73), Expect = 0.19
Identities = 34/140 (24%), Positives = 57/140 (40%), Gaps = 4/140 (2%)
Frame = +2
Query: 20 TRFTPTRLRTLRTVNAISKSKIPFNPND-PFLSKLASVAESSPETLFNPSSSPDNLPFLD 78
+ FT T+ +T S K NPN P L SSP+ NP +SP LP
Sbjct: 104 SHFTTTKWKTSGKERRHSPKKPQKNPNL*PPPPPLLESPISSPKPPRNPKNSPPKLPRKQ 283
Query: 79 IFDSPQLMATP---AQLERSASYSQQRRPKRPPPDLPSLLLNGRIVYIGMPLVPAVTELV 135
+ +P L P + ++ + P +PP + LL + + + L+
Sbjct: 284 MRSNPPLSVKPIRSSPFPITSPFLLNSPPLQPPQQVLP*LLRWK-------TLNDLVSLM 442
Query: 136 IAELMFLQWMAPKEPIYIYI 155
I + +F + P+ I++ I
Sbjct: 443 IFDRLFRDLLPPRSNIFLLI 502
>TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 805
Score = 30.8 bits (68), Expect = 0.73
Identities = 19/64 (29%), Positives = 30/64 (46%)
Frame = +3
Query: 8 PISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLFNP 67
P+S TS PAS+ +P T + + S +P + P A SSP + +P
Sbjct: 120 PVSTTTSPPASTPASSPVSTTTSPPASTPASSPVPTTTSPP-----APTPASSPVSTNSP 284
Query: 68 SSSP 71
++SP
Sbjct: 285 TASP 296
>TC78095 weakly similar to PIR|G84776|G84776 hypothetical protein At2g36090
[imported] - Arabidopsis thaliana, partial (19%)
Length = 1467
Score = 30.8 bits (68), Expect = 0.73
Identities = 26/97 (26%), Positives = 38/97 (38%), Gaps = 1/97 (1%)
Frame = +3
Query: 13 TSMPASSTRF-TPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLFNPSSSP 71
TS+P S+ F T ++ L + S + PN+ + + S T NP+SSP
Sbjct: 126 TSLPLSTLSFLTQHKIFILFLLKPFQYSNTNYQPNNIHIHLWIHQSPPSTPTYSNPTSSP 305
Query: 72 DNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPP 108
D L SP P SA + + PP
Sbjct: 306 D----LTAHHSPPSPPPPPTFTTSAQNTTYGKKSSPP 404
>TC82261 similar to GP|10177249|dbj|BAB10717. contains similarity to
phytocyanin/early nodulin-like protein~gene_id:K19P17.3,
partial (7%)
Length = 740
Score = 30.8 bits (68), Expect = 0.73
Identities = 22/58 (37%), Positives = 30/58 (50%)
Frame = +1
Query: 32 TVNAISKSKIPFNPNDPFLSKLASVAESSPETLFNPSSSPDNLPFLDIFDSPQLMATP 89
T + +S S+ P N FL L + E SP +PSSSP P I SP +++TP
Sbjct: 418 TPSPVSPSESPSNSPSFFLPSLLTSPELSP----SPSSSPSPSPV--ISPSPTVLSTP 573
>TC81337 similar to GP|15723293|gb|AAL06332.1 U2 auxiliary factor small
subunit {Arabidopsis thaliana}, partial (81%)
Length = 1190
Score = 29.6 bits (65), Expect = 1.6
Identities = 25/96 (26%), Positives = 35/96 (36%), Gaps = 22/96 (22%)
Frame = -2
Query: 44 NPNDPFLSKLASVAESSPETLFNPSSS-----PDNLPFLDIFDSPQLMATPAQLERSASY 98
NP PFL+ + SP T+ P+ S P ++P +P + R S+
Sbjct: 886 NPRSPFLTSFPCLPPISPPTISIPTISFPLPIPISIPNPIPITTPSSTRIIISITRPLSF 707
Query: 99 SQQRRPKRP-----------------PPDLPSLLLN 117
S P P PP LP+ LLN
Sbjct: 706 SASASPSPPRTITPAPAPAPLRRNQFPPQLPTNLLN 599
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,454,847
Number of Sequences: 36976
Number of extensions: 105509
Number of successful extensions: 789
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 772
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 779
length of query: 322
length of database: 9,014,727
effective HSP length: 96
effective length of query: 226
effective length of database: 5,465,031
effective search space: 1235097006
effective search space used: 1235097006
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144541.11


