

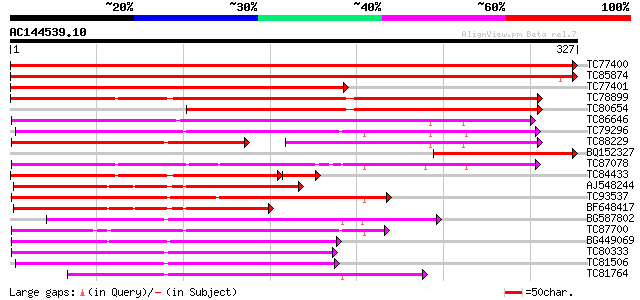
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144539.10 - phase: 0
(327 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77400 similar to GP|1247325|emb|CAA01814.1 beta-1 3-glucanase ... 665 0.0
TC85874 homologue to PIR|T09401|T09401 1 3-beta-glucanase (EC 3.... 513 e-146
TC77401 similar to GP|1247325|emb|CAA01814.1 beta-1 3-glucanase ... 369 e-103
TC78899 similar to GP|3900936|emb|CAA10167.1 glucan endo-1 3-bet... 293 6e-80
TC80654 similar to GP|3900936|emb|CAA10167.1 glucan endo-1 3-bet... 220 7e-58
TC86646 similar to PIR|E84471|E84471 probable beta-1 3-glucanase... 216 1e-56
TC79296 weakly similar to GP|14279169|gb|AAK58515.1 beta-1 3-glu... 195 2e-50
TC88229 similar to PIR|E84471|E84471 probable beta-1 3-glucanase... 110 2e-48
BQ152327 similar to GP|261212|gb|AA beta-1 3-glucanase {Pisum sa... 178 2e-45
TC87078 similar to PIR|T50645|T50645 glucan endo-1 3-beta-D-gluc... 177 4e-45
TC84433 similar to PIR|T05957|T05957 1 3-beta-glucanase (EC 3.2.... 157 1e-40
AJ548244 similar to GP|3900936|emb glucan endo-1 3-beta-d-glucos... 158 2e-39
TC93537 weakly similar to GP|6714534|dbj|BAA89481.1 beta-1 3-glu... 157 4e-39
BF648417 similar to PIR|T05959|T05 1 3-beta-glucanase (EC 3.2.1.... 142 2e-34
BG587802 similar to GP|20197543|gb putative beta-1 3-glucanase {... 141 4e-34
TC87700 weakly similar to PIR|S31196|S31196 hypothetical protein... 135 3e-32
BG449069 weakly similar to GP|21741726|em OSJNBa0073L04.6 {Oryza... 133 1e-31
TC80333 similar to GP|9758115|dbj|BAB08587.1 beta-1 3-glucanase-... 132 2e-31
TC81506 weakly similar to GP|22748323|gb|AAN05325.1 Putative bet... 125 2e-29
TC81764 similar to GP|17104805|gb|AAL34291.1 putative glucan end... 125 3e-29
>TC77400 similar to GP|1247325|emb|CAA01814.1 beta-1 3-glucanase {Glycine
max}, partial (88%)
Length = 1352
Score = 665 bits (1715), Expect = 0.0
Identities = 327/327 (100%), Positives = 327/327 (100%)
Frame = +3
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN
Sbjct: 222 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 401
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD
Sbjct: 402 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 581
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP
Sbjct: 582 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 761
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG
Sbjct: 762 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 941
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 300
GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ
Sbjct: 942 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 1121
Query: 301 KKYPFGFQGKIDGTNLFNVSFPLKSDI 327
KKYPFGFQGKIDGTNLFNVSFPLKSDI
Sbjct: 1122KKYPFGFQGKIDGTNLFNVSFPLKSDI 1202
>TC85874 homologue to PIR|T09401|T09401 1 3-beta-glucanase (EC 3.2.1.-)
acidic - alfalfa, complete
Length = 1336
Score = 513 bits (1322), Expect = e-146
Identities = 253/330 (76%), Positives = 284/330 (85%), Gaps = 3/330 (0%)
Frame = +1
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
MMGNNLP E I+L K+NNIKRMRLYDPNQAAL ALRNSGIEL+LGVPNSDLQ +ATN+
Sbjct: 178 MMGNNLPPANEVINLYKANNIKRMRLYDPNQAALNALRNSGIELILGVPNSDLQTLATNS 357
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
D A QWVQ+NVLNF+PSVKIKYIAVGNEV+PVGGSS A++VLPA QNIYQAIRA+ LHD
Sbjct: 358 DNARQWVQRNVLNFWPSVKIKYIAVGNEVSPVGGSSWLAQYVLPATQNIYQAIRAQGLHD 537
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
QIKVSTAIDMT+IG S+PPSKGSFR+DVRSYLDP IGYLVYA APL N+Y YFS+ NP
Sbjct: 538 QIKVSTAIDMTLIGNSFPPSKGSFRNDVRSYLDPFIGYLVYAGAPLLVNVYPYFSHVGNP 717
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
+DISL YALFTSP V+V DG GYQNLFDA+LDS+HAA+DNTGIG+V VVVSESGWPSDG
Sbjct: 718 RDISLPYALFTSPGVMVQDGPNGYQNLFDAMLDSVHAALDNTGIGWVNVVVSESGWPSDG 897
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 300
G AT+YDNAR+YLDNLIRHV GTP R ETYIF +FDENQK+PELEKHFGVFYPNKQ
Sbjct: 898 GAATSYDNARIYLDNLIRHVGKGTPRRPWATETYIFAMFDENQKSPELEKHFGVFYPNKQ 1077
Query: 301 KKYPFGFQGKIDGTNL---FNVSFPLKSDI 327
KKYPFGF G+ +G + FN + LKSD+
Sbjct: 1078KKYPFGFGGERNGEIVNGDFNATISLKSDM 1167
>TC77401 similar to GP|1247325|emb|CAA01814.1 beta-1 3-glucanase {Glycine
max}, partial (59%)
Length = 737
Score = 369 bits (948), Expect = e-103
Identities = 186/195 (95%), Positives = 186/195 (95%)
Frame = +2
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
M G NLPSQREAIDLCKSNNIKRMRLYDPN ALEALRNSGIELMLGVPNSDLQNIA N
Sbjct: 152 MEGTNLPSQREAIDLCKSNNIKRMRLYDPNPDALEALRNSGIELMLGVPNSDLQNIANNK 331
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
DIA QWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKN D
Sbjct: 332 DIANQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNFQD 511
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP
Sbjct: 512 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 691
Query: 181 KDISLQYALFTSPNV 195
KDISLQYALFTSPNV
Sbjct: 692 KDISLQYALFTSPNV 736
>TC78899 similar to GP|3900936|emb|CAA10167.1 glucan endo-1
3-beta-d-glucosidase {Cicer arietinum}, partial (94%)
Length = 1296
Score = 293 bits (750), Expect = 6e-80
Identities = 154/308 (50%), Positives = 204/308 (66%), Gaps = 1/308 (0%)
Frame = +3
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
M+GNNLPS+++ ++L KS I +MR++ P++ AL+ALR S IEL+L V L ++ N
Sbjct: 147 MIGNNLPSRQDVVNLYKSRGINQMRIFFPDEPALQALRGSNIELILDVAKETLPSLRNAN 326
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
+ A WV K V + +VKIKYI+VGNE+ P + A+++LPA+QNI AI + NL
Sbjct: 327 E-ATNWVNKYVRPYAQNVKIKYISVGNEIKP---NDNEAQYILPAMQNIQNAISSANLQG 494
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
QIKVSTAIDMT+IG S+PP+ G F + Y+ PII +L APL AN+Y YF+Y +
Sbjct: 495 QIKVSTAIDMTLIGKSFPPNDGVFSDQAKPYIQPIINFLNNNGAPLLANVYPYFAYIGDK 674
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
+I L YALF + + GYQNLFDA LDS++AA++ G VK+VVSESGWPS
Sbjct: 675 VNIPLDYALFRQQG----NNAVGYQNLFDAQLDSVYAALEKVGASGVKIVVSESGWPSAA 842
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQK-NPELEKHFGVFYPNK 299
G + + DNA Y NLI HVK GTP R G IETY+F +FDENQK E+HFG+F P+K
Sbjct: 843 GDSASTDNAATYYRNLINHVKNGTPKRPGAIETYLFAMFDENQKTGAATEQHFGLFNPDK 1022
Query: 300 QKKYPFGF 307
KY F
Sbjct: 1023SPKYQISF 1046
>TC80654 similar to GP|3900936|emb|CAA10167.1 glucan endo-1
3-beta-d-glucosidase {Cicer arietinum}, partial (56%)
Length = 746
Score = 220 bits (560), Expect = 7e-58
Identities = 112/206 (54%), Positives = 137/206 (66%), Gaps = 1/206 (0%)
Frame = +3
Query: 103 LPAIQNIYQAIRAKNLHDQIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYA 162
LPA+QNI AI A NL QIKVS AIDMT+IG SYPP+ G F + Y+ PII +L
Sbjct: 9 LPAMQNIQNAISAANLQGQIKVSIAIDMTLIGNSYPPNNGVFTDQAKPYIQPIINFLKNN 188
Query: 163 NAPLFANIYSYFSYKDNPKDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNT 222
APL AN+Y YF+Y +N + ISL YALF + GY+NLFDA LDS++AA++
Sbjct: 189 GAPLLANVYPYFAYINNKQSISLDYALFRQQG----NNQVGYRNLFDAQLDSVYAALEKV 356
Query: 223 GIGFVKVVVSESGWPSDGGFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDEN 282
G VK+VVSESGWPS GG + + DNA Y NLI HV+ GTP R G IETY+F +FDEN
Sbjct: 357 GASGVKIVVSESGWPSAGGDSASTDNAATYYRNLINHVRNGTPKRPGAIETYLFAMFDEN 536
Query: 283 QK-NPELEKHFGVFYPNKQKKYPFGF 307
QK E+HFG+F PN+ KY F
Sbjct: 537 QKTGAATEQHFGLFNPNRTPKYQISF 614
>TC86646 similar to PIR|E84471|E84471 probable beta-1 3-glucanase [imported]
- Arabidopsis thaliana, partial (73%)
Length = 1257
Score = 216 bits (549), Expect = 1e-56
Identities = 120/311 (38%), Positives = 188/311 (59%), Gaps = 9/311 (2%)
Frame = +2
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+ NNLPS + + L KS I +++YD + + L AL S I+L + +PN L A +
Sbjct: 167 IANNLPSATKVVQLLKSQGITHVKIYDTDPSVLRALSGSKIKLTVDLPNQQLFAAAKSQS 346
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQ 121
A+ WV++N++ + P+ I+ IAVGNEV +S K+++PA++NIY++++ NLH+
Sbjct: 347 FALSWVERNIVAYQPNTIIEAIAVGNEVFVDPNNS--TKYLVPAMKNIYRSLQKHNLHND 520
Query: 122 IKVSTAIDMTMIGTSYPPSKGSFRSD-VRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
IKVS+ I ++ +G SYP S GSFR + ++ P++ +L + L N+Y +F+Y+ N
Sbjct: 521 IKVSSPIALSALGNSYPSSSGSFRPELIQPVFKPMLDFLRETGSYLMVNVYPFFAYESNA 700
Query: 181 KDISLQYALF-TSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSD 239
ISL YALF +P V Y NLFDA +D++ AA+ + VVVSE+GWPS
Sbjct: 701 DVISLDYALFRENPGQVDAGNGLRYLNLFDAQIDAVFAALSRLKYDDINVVVSETGWPSK 880
Query: 240 GG---FATTYDNARVYLDNLIRHV--KGGTPMR-SGPIETYIFGLFDENQK-NPELEKHF 292
G + +NA Y NL+R + GTP+R + ++F LF+ENQK P E++F
Sbjct: 881 GDGNEVGASVENAAAYNANLVRKILTSKGTPLRPKADLTVFLFALFNENQKPGPTSERNF 1060
Query: 293 GVFYPNKQKKY 303
G+FYP+++K Y
Sbjct: 1061GLFYPDEKKVY 1093
>TC79296 weakly similar to GP|14279169|gb|AAK58515.1 beta-1 3-glucanase-like
protein {Olea europaea}, partial (90%)
Length = 1710
Score = 195 bits (496), Expect = 2e-50
Identities = 109/312 (34%), Positives = 182/312 (57%), Gaps = 9/312 (2%)
Frame = +3
Query: 4 NNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNNDIA 63
+NLP L +S I ++R+Y + A +++L NSGI +++G N+D+ ++A++ + A
Sbjct: 207 DNLPPPEATAKLLQSTTIGKVRIYGADPAIIKSLANSGIGIVIGAANNDIPSLASDPNAA 386
Query: 64 IQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQIK 123
QW+ NVL +YP+ I I VGNEV G ++ ++PAI+N+ A+ + L ++K
Sbjct: 387 TQWINTNVLPYYPASNITLITVGNEVLNSGDEGLVSQ-LMPAIRNVQTALSSVKLGGKVK 563
Query: 124 VSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNPKDI 183
V+T M ++ S PPS GSF +R+ L+ ++ +L +P N Y +F+Y+ +P+
Sbjct: 564 VTTVHSMAVLAQSDPPSSGSFNPALRNTLNQLLAFLKDNKSPFTVNPYPFFAYQSDPRPE 743
Query: 184 SLQYALFTSPNVVVWDGSRG--YQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDGG 241
+L + LF PN D G Y N+FDA +D++H+A+ +++VV+E+GWPS G
Sbjct: 744 TLTFCLF-QPNSGRVDSGNGKLYTNMFDAQVDAVHSALSAMSYEDIEIVVAETGWPSSGD 920
Query: 242 ---FATTYDNARVYLDNLIRHVKG--GTPMRSG-PIETYIFGLFDENQK-NPELEKHFGV 294
+ +NA+ Y NLI H++ GTP+ G ++TYIF L+DE+ K P E+ FG+
Sbjct: 921 NNEVGPSVENAKAYNGNLITHLRSLVGTPLIPGKSVDTYIFALYDEDLKPGPGSERAFGL 1100
Query: 295 FYPNKQKKYPFG 306
F + Y G
Sbjct: 1101FKTDLSMSYDIG 1136
>TC88229 similar to PIR|E84471|E84471 probable beta-1 3-glucanase [imported]
- Arabidopsis thaliana, partial (69%)
Length = 1368
Score = 110 bits (275), Expect(2) = 2e-48
Identities = 55/137 (40%), Positives = 91/137 (66%)
Frame = +1
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+ NNLPS + + L KS I R++LYD + A L++L SGI++ + +PN L + A
Sbjct: 127 IANNLPSAFKVVKLLKSQGIDRVKLYDTDPAVLKSLSGSGIKVTVNLPNEQLFHTARKLS 306
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQ 121
A+ W+QKNV+ ++P +I+ IAVGNEV + K+++PA++NI++A+ NLH+
Sbjct: 307 YALTWLQKNVVVYHPKTQIEAIAVGNEV--FVDTHNTTKYLIPAMKNIHKALVKFNLHNS 480
Query: 122 IKVSTAIDMTMIGTSYP 138
IK+S+ I ++ +G+SYP
Sbjct: 481 IKISSPIALSALGSSYP 531
Score = 99.8 bits (247), Expect(2) = 2e-48
Identities = 61/156 (39%), Positives = 90/156 (57%), Gaps = 8/156 (5%)
Frame = +3
Query: 160 VYANAPLFANIYSYFSYKDNPKDISLQYALFT-SPNVVVWDGSRGYQNLFDALLDSLHAA 218
V ++ L N+Y +F+Y+ N ISL YALF +P V Y N+FDA +D++ AA
Sbjct: 600 VKTSSYLMVNVYPFFAYESNADVISLNYALFRENPGNVDPGNGLKYYNIFDAQIDAVFAA 779
Query: 219 IDNTGIGFVKVVVSESGWPSDGG---FATTYDNARVYLDNLIRHV--KGGTPMR-SGPIE 272
++ V+VVVSE+GWPS G + NA Y NL++ + GGTP+R + +
Sbjct: 780 LNVLQYDDVRVVVSETGWPSKGDSNEVGASPQNAAAYNGNLVKKILNNGGTPLRPNANLT 959
Query: 273 TYIFGLFDENQK-NPELEKHFGVFYPNKQKKYPFGF 307
Y+F LF+EN K E++FG+FYP+ +K Y F
Sbjct: 960 VYLFALFNENGKVGLTSERNFGMFYPDMKKVYDVPF 1067
>BQ152327 similar to GP|261212|gb|AA beta-1 3-glucanase {Pisum sativum},
partial (18%)
Length = 387
Score = 178 bits (452), Expect = 2e-45
Identities = 83/83 (100%), Positives = 83/83 (100%)
Frame = +3
Query: 245 TYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQKKYP 304
TYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQKKYP
Sbjct: 39 TYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQKKYP 218
Query: 305 FGFQGKIDGTNLFNVSFPLKSDI 327
FGFQGKIDGTNLFNVSFPLKSDI
Sbjct: 219 FGFQGKIDGTNLFNVSFPLKSDI 287
>TC87078 similar to PIR|T50645|T50645 glucan endo-1 3-beta-D-glucosidase (EC
3.2.1.39) [imported] - garden pea, partial (94%)
Length = 1628
Score = 177 bits (450), Expect = 4e-45
Identities = 116/315 (36%), Positives = 181/315 (56%), Gaps = 10/315 (3%)
Frame = +1
Query: 2 MGNNLPSQREAIDLCKSNNI-KRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
+ +NLP + K+N I R++++D + L+A N+GI + + PN D+ + N
Sbjct: 106 LADNLPPPATVANFLKTNTIVDRVKIFDVSPQILQAFANTGISVTVTAPNGDIAALGNIN 285
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
A QWVQ+ + FYP+ KI YI VG+EV G + V PA++ ++ A+ A+ ++D
Sbjct: 286 S-ARQWVQQKIKPFYPATKINYILVGSEVLHWGDGNMIRGLV-PAMRTLHSALVAEGIND 459
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSY-LDPIIGYLVYANAPLFANIYSYFSYKDN 179
IKV+TA + ++ S PPS G FR + + P++ +L P N Y YF Y N
Sbjct: 460 -IKVTTAHSLIIMRQSLPPSAGKFRPGFAKHVIAPMLKFLRETRTPFMVNPYPYFGY--N 630
Query: 180 PKDISLQYALFTSPNVVVWDGSRG--YQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWP 237
PK+++ +ALF PN ++D + Y N FDAL+D++++A+ G G V + V E+GWP
Sbjct: 631 PKNVN--FALFR-PNRGLFDRNTRLTYTNQFDALMDAVYSAMKGLGFGDVDIAVGETGWP 801
Query: 238 S--DGGFATTYDNARVYLDNLIRHVKG--GTP-MRSGPIETYIFGLFDENQK-NPELEKH 291
S DG A + NA+ Y LIRH++ GTP M + ET++F LF+ENQK P E++
Sbjct: 802 SVCDGWDACSVANAQSYNGELIRHLEAGRGTPLMPNRRFETFLFALFNENQKPGPIAERN 981
Query: 292 FGVFYPNKQKKYPFG 306
+G+F P+ Y G
Sbjct: 982 WGLFRPDFSPVYDSG 1026
>TC84433 similar to PIR|T05957|T05957 1 3-beta-glucanase (EC 3.2.1.-) Glu3 -
soybean (fragment), partial (72%)
Length = 682
Score = 157 bits (397), Expect(2) = 1e-40
Identities = 81/157 (51%), Positives = 107/157 (67%)
Frame = +1
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
++GNNLPS +E +DL KSN I +MR+Y P++ AL+AL+ S IEL+L V L ++ N
Sbjct: 148 VLGNNLPSSQEVVDLYKSNGIDKMRIYFPDEQALQALKGSNIELILDVAKETLSSLTDGN 327
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
+ A WVQK V + VKIKYI VGNE+ P + A+++ A+QNI AI + NL
Sbjct: 328 E-ATNWVQKYVTPYAQDVKIKYITVGNEIKP---NDNEAQYIATAMQNIQNAISSANLQG 495
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIG 157
QIKVSTAIDMT+IGTSYPP+ G+F + YL PIIG
Sbjct: 496 QIKVSTAIDMTLIGTSYPPNDGAFTDQAKQYLQPIIG 606
Score = 26.9 bits (58), Expect(2) = 1e-40
Identities = 11/22 (50%), Positives = 14/22 (63%)
Frame = +2
Query: 158 YLVYANAPLFANIYSYFSYKDN 179
+L APL AN+Y YF+Y N
Sbjct: 608 FLKNNGAPLLANVYPYFAYIGN 673
>AJ548244 similar to GP|3900936|emb glucan endo-1 3-beta-d-glucosidase {Cicer
arietinum}, partial (46%)
Length = 649
Score = 158 bits (400), Expect = 2e-39
Identities = 87/167 (52%), Positives = 117/167 (69%)
Frame = -3
Query: 3 GNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNNDI 62
GNNLP+++ +DL KS I ++R+Y+P++ L+ALR+S IE++LGVPN+ L+++ TN
Sbjct: 488 GNNLPTKKAVVDLYKSKGIGKIRIYNPDEGILQALRSSNIEVILGVPNNVLKSL-TNAQT 312
Query: 63 AIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQI 122
A WV K V Y VKIKYIAVGNEV+P GS++ + VLPA+QNI +AI + NL QI
Sbjct: 311 ASDWVNKYV-KAYSIVKIKYIAVGNEVHP--GSAESSS-VLPAMQNIQKAISSANLQGQI 144
Query: 123 KVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFAN 169
KVSTAID T+IG SYPP G F Y+ PI+ +LV +PL AN
Sbjct: 143 KVSTAIDTTLIGKSYPPKDGVFSDAASGYIKPIVNFLVSNGSPLLAN 3
>TC93537 weakly similar to GP|6714534|dbj|BAA89481.1 beta-1 3-glucanase
{Salix gilgiana}, partial (46%)
Length = 736
Score = 157 bits (398), Expect = 4e-39
Identities = 79/222 (35%), Positives = 142/222 (63%), Gaps = 3/222 (1%)
Frame = +1
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+GNNLP+ ++ L K+ KR+++YD N L+AL N+GI++ + +PN + N+++N
Sbjct: 76 LGNNLPTPTTSVSLIKNLKAKRVKIYDANPQILKALENTGIQVSIMLPNELVTNVSSNQT 255
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQ 121
+A QWVQ N++ FY I+Y+ VGNE+ ++Q ++PA+ + ++ LH +
Sbjct: 256 LANQWVQTNLVPFYSKTLIRYLLVGNELIS-STTNQTWPHIVPAMYRMKHSLTIFGLH-K 429
Query: 122 IKVSTAIDMTMIGTSYPPSKGSFRSDVR-SYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
+KV T + M ++ TS+PPS G+FR+D+ S + P++ +L N+ F ++Y +F++ +P
Sbjct: 430 VKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMVEFLHVTNSFFFLDVYPFFAWTSDP 609
Query: 181 KDISLQYALFTSPNVVVWDGSRG--YQNLFDALLDSLHAAID 220
+I+L YALF S N+ V D G Y NLF+ ++D+++ A++
Sbjct: 610 ININLDYALFESDNITVTDSGTGLVYTNLFEQMVDAVYFAME 735
>BF648417 similar to PIR|T05959|T05 1 3-beta-glucanase (EC 3.2.1.-) Glu5 -
soybean (fragment), partial (63%)
Length = 657
Score = 142 bits (358), Expect = 2e-34
Identities = 79/150 (52%), Positives = 106/150 (70%)
Frame = +1
Query: 3 GNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNNDI 62
GNNLP+++ +DL KS I ++R+Y+P++ L+ALR+S IE++LGVPN+ L+++ TN
Sbjct: 220 GNNLPTKKAVVDLYKSKGIGKIRIYNPDEGILQALRSSNIEVILGVPNNVLKSL-TNAQT 396
Query: 63 AIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQI 122
A WV K V Y VKIKYIAVGNEV+P GS++ + VLPA+QNI +AI + NL QI
Sbjct: 397 ASDWVNKYV-KAYSIVKIKYIAVGNEVHP--GSAESSS-VLPAMQNIQKAISSANLQGQI 564
Query: 123 KVSTAIDMTMIGTSYPPSKGSFRSDVRSYL 152
KVSTAID T+IG SYPP G F Y+
Sbjct: 565 KVSTAIDTTLIGKSYPPKDGVFSDAASGYI 654
>BG587802 similar to GP|20197543|gb putative beta-1 3-glucanase {Arabidopsis
thaliana}, partial (48%)
Length = 753
Score = 141 bits (355), Expect = 4e-34
Identities = 83/232 (35%), Positives = 129/232 (54%), Gaps = 4/232 (1%)
Frame = +1
Query: 22 KRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNNDIAIQWVQKNVLNFYPSVKIK 81
+ +RL+D +++ L AL +GI++++ VPN +L I +N A WV +NV+ PS I
Sbjct: 1 QHVRLFDADKSMLLALAKTGIQVIVSVPNDELLGIGQSNATAANWVARNVIAHVPSTNIT 180
Query: 82 YIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQIKVSTAIDMTMIGTSYPPSK 141
IAVG+EV + A ++ A+Q I A+ A NL DQIKVST + ++I S+PPS+
Sbjct: 181 AIAVGSEV--LTSLPNAAPVLVSALQFIQSALVAANLDDQIKVSTPLSTSIILDSFPPSQ 354
Query: 142 GSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNPKDISLQYALF--TSPNVVVWD 199
F + P++ +L + L N+Y Y+ Y + I L YALF PN D
Sbjct: 355 AFFNRTWDPVMSPLLKFLQSTGSYLMLNVYPYYDYMQSNDVIPLDYALFRPLPPNKEAVD 534
Query: 200 GSR--GYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDGGFATTYDNA 249
+ Y N+FDA++D+ + A+ + ++V+ESGWPS GGF N+
Sbjct: 535 SNTLLHYTNVFDAVIDAAYFAMSYLKFTNIPILVTESGWPSKGGFVRAGRNS 690
>TC87700 weakly similar to PIR|S31196|S31196 hypothetical protein - potato,
partial (47%)
Length = 791
Score = 135 bits (339), Expect = 3e-32
Identities = 79/221 (35%), Positives = 124/221 (55%), Gaps = 3/221 (1%)
Frame = +3
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+ NNLPS L KS N+ R++LYD + L NS +E M+G+ +DLQ++ +
Sbjct: 144 IANNLPSHSRVAVLIKSLNVSRIKLYDADPNVLTTFSNSNVEFMIGL--NDLQSM--KDP 311
Query: 62 IAIQ-WVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
I Q WVQ+NV + P KI I VGNEV + + +LPA++++Y A+ L
Sbjct: 312 IKAQSWVQQNVQPYLPQTKITSINVGNEVLGNNDINSYNN-LLPAMKSVYNALVNLGLSQ 488
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
Q+ V+T+ ++ S+PPS G+FR D+ Y+ P++ + +P N Y +F+YK +P
Sbjct: 489 QVTVTTSHSFIIMSNSFPPSSGAFREDLIQYIQPLLSFQAQIKSPFLINAYPFFAYKGDP 668
Query: 181 KDISLQYALFTSPNVVVWDGSRG--YQNLFDALLDSLHAAI 219
+ +SL Y LF PN D + Y N+ A +D+ +AAI
Sbjct: 669 QHVSLNYVLF-QPNAGSIDPATNLHYDNMLYAQIDARYAAI 788
>BG449069 weakly similar to GP|21741726|em OSJNBa0073L04.6 {Oryza sativa},
partial (46%)
Length = 668
Score = 133 bits (334), Expect = 1e-31
Identities = 72/190 (37%), Positives = 106/190 (54%)
Frame = +2
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+ NLPS L +S N+ R++LYD + L A S +E ++G+ N LQN+ TN
Sbjct: 62 IARNLPSHSRVAMLIRSMNVTRIKLYDADPNVLLAFSKSNVEFVIGIGNEHLQNM-TNPS 238
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQ 121
A W+Q +VL + KI I VGNEV +Q +LP++QN++ ++ L
Sbjct: 239 KAQNWIQHHVLPYLSQTKINCITVGNEVFN-SNDTQLILNLLPSMQNVHNSLVKLGLDQV 415
Query: 122 IKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNPK 181
I V+TA ++ SYPPS GSFRSD+ Y+ PI+ +L + N Y +F+YKDNP
Sbjct: 416 ITVTTAHSFNILDNSYPPSSGSFRSDLIQYIQPIVEFLDEIKSSFHINAYPFFAYKDNPN 595
Query: 182 DISLQYALFT 191
++SL FT
Sbjct: 596 EVSLTMHYFT 625
>TC80333 similar to GP|9758115|dbj|BAB08587.1 beta-1 3-glucanase-like
protein {Arabidopsis thaliana}, partial (46%)
Length = 836
Score = 132 bits (331), Expect = 2e-31
Identities = 66/189 (34%), Positives = 114/189 (59%), Gaps = 1/189 (0%)
Frame = +2
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+ +NLP+ + ++L K+ R++LYD + L AL NSGI++ + +PN L + A +
Sbjct: 134 IADNLPTPSKVVELLKAQGFNRVKLYDTDATVLTALANSGIKVTVAMPNELLSSAAADQS 313
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQ 121
W+Q N+LN YP+ +I+ IAVGNEV +++PA++N++ +++ +NL Q
Sbjct: 314 YTDTWIQSNILNHYPATEIEAIAVGNEV--FVDPKNTTNYLVPAMKNVHASLQKQNLDKQ 487
Query: 122 IKVSTAIDMTMIGTSYPPSKGSFRSD-VRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
I +S+ I ++ + +SYP S GSF+++ V + P++ +L + L N Y +F+Y N
Sbjct: 488 ILISSPIALSALQSSYPTSTGSFKTELVEPVIKPMLEFLSQTGSYLMVNAYPFFAYAANS 667
Query: 181 KDISLQYAL 189
ISL YAL
Sbjct: 668 DTISLDYAL 694
>TC81506 weakly similar to GP|22748323|gb|AAN05325.1 Putative beta-1
3-glucanase {Oryza sativa (japonica cultivar-group)},
partial (39%)
Length = 850
Score = 125 bits (315), Expect = 2e-29
Identities = 70/187 (37%), Positives = 109/187 (57%)
Frame = +2
Query: 4 NNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNNDIA 63
+++P + + L K+ I+ +RLYD +QA L AL +GI++++ VPN + I +N A
Sbjct: 275 SDMPHPTQVVALLKAQKIQNVRLYDADQAMLVALAKTGIQVVITVPNEQILAIGQSNASA 454
Query: 64 IQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQIK 123
WV +NV+ YP+ I I VG+EV + AK ++ AI+ I+ A+ A NL Q+K
Sbjct: 455 ANWVSRNVVAHYPATNITAICVGSEV--LTTLPNVAKVLVNAIKYIHSALVASNLDRQVK 628
Query: 124 VSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNPKDI 183
VST + ++I S+PPS+ F + S L PI+ +L ++ L NIY Y+ Y + I
Sbjct: 629 VSTPLPSSIILDSFPPSQAFFNRSLNSVLIPILDFLQSTDSYLMLNIYPYYDYMQSNGVI 808
Query: 184 SLQYALF 190
L YALF
Sbjct: 809 XLXYALF 829
>TC81764 similar to GP|17104805|gb|AAL34291.1 putative glucan endo-1
3-beta-glucosidase precursor {Arabidopsis thaliana},
partial (71%)
Length = 1560
Score = 125 bits (313), Expect = 3e-29
Identities = 71/212 (33%), Positives = 118/212 (55%), Gaps = 4/212 (1%)
Frame = +2
Query: 34 LEALRNSGIELMLGVPNSDLQNIATNNDIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVG 93
L+AL + I++M+GV N ++ I + A W+ KNV+ + PS I IAVG+EV +
Sbjct: 35 LQALSKTNIDVMVGVTNEEVLRIGESPSAAAAWINKNVVAYVPSTNITAIAVGSEV--LS 208
Query: 94 GSSQFAKFVLPAIQNIYQAIRAKNLHDQIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLD 153
A ++PA+ ++++A+ A NL+ ++KVST M +I +PPS +F S S +
Sbjct: 209 TIPNVAPVLVPAMNSLHKALVAANLNFRVKVSTPQSMDIIPKPFPPSTATFNSSWNSTIY 388
Query: 154 PIIGYLVYANAPLFANIYSYFSYKDNPKDISLQYALF----TSPNVVVWDGSRGYQNLFD 209
++ +L N+ N Y Y+ Y L+YALF + +V + Y ++FD
Sbjct: 389 QVLQFLRNTNSSFMLNAYPYYGYTKGDGIFPLEYALFRPLPSVKQIVDPNTLYHYNSMFD 568
Query: 210 ALLDSLHAAIDNTGIGFVKVVVSESGWPSDGG 241
A++D+ + +ID + VVV+E+GWPS GG
Sbjct: 569 AMVDATYYSIDALNFKDIPVVVTETGWPSFGG 664
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,875,163
Number of Sequences: 36976
Number of extensions: 111571
Number of successful extensions: 585
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 537
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 537
length of query: 327
length of database: 9,014,727
effective HSP length: 96
effective length of query: 231
effective length of database: 5,465,031
effective search space: 1262422161
effective search space used: 1262422161
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144539.10


