

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144517.16 - phase: 0
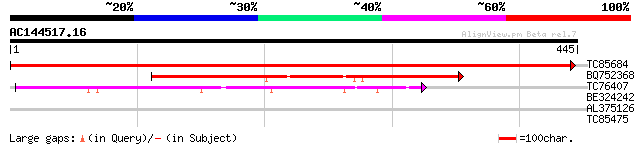
(445 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85684 homologue to GP|3175990|emb|CAA06731.1 GDP dissociation ... 751 0.0
BQ752368 similar to PIR|T38215|T38 probable secretory pathway GD... 264 6e-71
TC76407 weakly similar to GP|20453227|gb|AAM19852.1 AT3g06540/F5... 115 3e-26
BE324242 weakly similar to GP|12322677|gb| Rab escort protein p... 31 0.84
AL375126 homologue to GP|3175990|emb| GDP dissociation inhibitor... 31 1.1
TC85475 homologue to PIR|T06507|T06507 reversibly glycosylatable... 28 5.5
>TC85684 homologue to GP|3175990|emb|CAA06731.1 GDP dissociation inhibitor
{Cicer arietinum}, complete
Length = 1849
Score = 751 bits (1938), Expect = 0.0
Identities = 360/444 (81%), Positives = 405/444 (91%)
Frame = +2
Query: 1 MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDQNDYYGGASTSLNLTQLFKRYRGDDK 60
MDEEYDVIVLGTGLKEC+LSGLLSVDGLKVLHMD+NDYYGG STSLNL QL+K++RGDDK
Sbjct: 137 MDEEYDVIVLGTGLKECVLSGLLSVDGLKVLHMDRNDYYGGESTSLNLIQLWKKFRGDDK 316
Query: 61 PPEELGSSREYNVDMIPKFMMANGALVRVLIHTDVTKYLNFKAVDGSFVYNKGKIYKVPA 120
PP LGSS++YN+DM PKF+MANG LVRVLIHTDVTKYL FKAVDGSFV+NK K++KVP+
Sbjct: 317 PPPHLGSSKDYNIDMNPKFIMANGTLVRVLIHTDVTKYLYFKAVDGSFVFNKAKVHKVPS 496
Query: 121 TDVEALKSPLMGLFEKRRARKFFIYVQDYEANDPKSHEGLDLNQVTARQLISKYGLEDDT 180
D+EALKSPLMG+FEKRRARKFFIYVQDY +DPK+H+G+DL +VT ++LI+K+GL+D+T
Sbjct: 497 NDMEALKSPLMGIFEKRRARKFFIYVQDYNESDPKTHDGMDLTRVTTKELIAKFGLDDNT 676
Query: 181 VDFIGHALALHLDDSYLDKPAKDFVDRVKTYAESLARFQGGSPYIYPLYGLGELPQAFAR 240
VDFIGHALALH DD YL++PA D V R+K YAESLARFQGGSPYIYPLYGLGELPQAFAR
Sbjct: 677 VDFIGHALALHRDDRYLNEPALDTVKRMKLYAESLARFQGGSPYIYPLYGLGELPQAFAR 856
Query: 241 LSAVYGGTYMLNKPECKVEFDGDGKAIGVTSDGETAKCKKVVCDPSYLPDKVQNVGKVAR 300
LSAVYGGTYMLNKPECKVEFD GK IGVTS+GETAKCKK+VCDPSYL +KV+ VGKVAR
Sbjct: 857 LSAVYGGTYMLNKPECKVEFDDQGKVIGVTSEGETAKCKKLVCDPSYLTNKVRKVGKVAR 1036
Query: 301 AICIMSHPIPDTNDSHSAQVILPQKQLGRKSDMYLFCCSYAHNVAPKGKYIAFVTTEAET 360
AI IMSHPIP+TNDS S QVILPQKQLGRKSDMYLFCCSY+HNV PKGK+IAFV+TEAET
Sbjct: 1037AIAIMSHPIPNTNDSQSVQVILPQKQLGRKSDMYLFCCSYSHNVCPKGKFIAFVSTEAET 1216
Query: 361 DQPQVELKPGIDLLGPVDEIFYDIYDRFEPTNDHATDGCFISTSYDPTTHFETTVKDVVQ 420
DQP +ELKPGIDLLGPVDEIF+D+YDRFEP N+ D CFISTSYD TTHFE+TV DV+
Sbjct: 1217DQPAIELKPGIDLLGPVDEIFFDMYDRFEPVNEPTLDNCFISTSYDATTHFESTVLDVLN 1396
Query: 421 MYSKITGKVLDLSVDLSAASAAAE 444
MY+ ITGKVLDL+VDLSAASAA E
Sbjct: 1397MYTLITGKVLDLTVDLSAASAAEE 1468
>BQ752368 similar to PIR|T38215|T38 probable secretory pathway GDP
dissociation inhibitor - fission yeast
(Schizosaccharomyces pombe), partial (44%)
Length = 811
Score = 264 bits (674), Expect = 6e-71
Identities = 134/254 (52%), Positives = 181/254 (70%), Gaps = 9/254 (3%)
Frame = +2
Query: 112 KGKIYKVPATDVEALKSPLMGLFEKRRARKFFIYVQDYEANDPKSHEGLDLNQVTARQLI 171
K + KVP+ EAL+SPLMG+FEKRR + F ++ +E D +H+GLD+N VT +++
Sbjct: 11 KATVAKVPSDAGEALRSPLMGIFEKRRMKSFIEWIGSFERKDTATHKGLDINNVTMKEVY 190
Query: 172 SKYGLEDDTVDFIGHALALHLDDSYLDKP--AKDFVDRVKTYAESLARFQGGSPYIYPLY 229
K+GLE T DFIGHA+AL D Y++KP A + ++R++ Y S+AR+ G SPYIYPLY
Sbjct: 191 DKFGLETGTRDFIGHAMALFPTDDYINKPGAAPEAIERIRLYGTSVARY-GKSPYIYPLY 367
Query: 230 GLGELPQAFARLSAVYGGTYMLNKPECKVEFDGDGKAIGV--TSDGET-----AKCKKVV 282
GLGELPQ FARLSA+YGGTYML+ +V ++G GKA+G+ T G K K ++
Sbjct: 368 GLGELPQGFARLSAIYGGTYMLHTNVDEVTYEG-GKAVGIKATMSGVEEMKFETKAKMII 544
Query: 283 CDPSYLPDKVQNVGKVARAICIMSHPIPDTNDSHSAQVILPQKQLGRKSDMYLFCCSYAH 342
DPSY P KV+ VG+V RAICI+ HP+ TNDS S+Q+I+PQ Q+GRK+D+Y+ C S AH
Sbjct: 545 GDPSYFPGKVKVVGQVLRAICILKHPLAGTNDSDSSQLIIPQSQIGRKNDIYIACVSSAH 724
Query: 343 NVAPKGKYIAFVTT 356
NV PKG +IA V+T
Sbjct: 725 NVCPKGYWIAIVST 766
>TC76407 weakly similar to GP|20453227|gb|AAM19852.1 AT3g06540/F5E6_13
{Arabidopsis thaliana}, partial (62%)
Length = 2233
Score = 115 bits (288), Expect = 3e-26
Identities = 104/393 (26%), Positives = 165/393 (41%), Gaps = 70/393 (17%)
Frame = +3
Query: 5 YDVIVLGTGLKECILSGLLSVDGLKVLHMDQNDYYGGASTSLNLTQLFKRYRGDDK---- 60
+D+I++GTGL E I+S S G +LH+D N +YG SL+ L K
Sbjct: 126 FDLIIVGTGLSESIISAAASAVGKTILHLDPNPFYGSHFASLSFEDLISHLNSPPKYITA 305
Query: 61 ----------------------PPEELGS-------------SREYNVDMI-PKFMMANG 84
E S SR++N+D+ P+ + +
Sbjct: 306 TATATTDNSDYTTVNLIQQTIFSDAEFNSCNSIEESQFLRENSRKFNLDLAGPRALFSAD 485
Query: 85 ALVRVLIHTDVTKYLNFKAVDGSFVYNKGK-IYKVPATDVEALKSPLMGLFEKRRARKFF 143
+ +L+ + +YL FK +D S VYN + + VP + + + L EK + KFF
Sbjct: 486 KTIDLLLKSGAAQYLEFKGIDASLVYNTNEGLVNVPDSRGAIFRDKNLSLKEKNQLMKFF 665
Query: 144 IYVQDY---EANDPKSHEGLDLNQVTARQLISKYGLEDDTVDFIGHALALHLDDSYLDKP 200
VQ + ++ S E ++ + V+ + K GL + +A+A+ D
Sbjct: 666 KLVQQHLGGNEDEKISEEDMESSFVS---YLEKLGLPSKIKAILLYAIAMVDFDQENGGV 836
Query: 201 AKDF------VDRVKTYAESLARFQGG-SPYIYPLYGLGELPQAFARLSAVYGGTYMLNK 253
KD +DR+ Y+ S+ R+ +YP+YG GELPQAF R +AV G Y+L
Sbjct: 837 CKDLLKTKDGIDRLAQYSSSVGRYPNAPGALLYPIYGEGELPQAFCRRAAVKGCIYVLRM 1016
Query: 254 PECKVEFD---GDGKAIGVTSDGETAKCKKVVCDPSY----------------LPDKVQN 294
P + D G K + ++S G+ K++ DPS+ + V
Sbjct: 1017PVISLLIDKVTGSYKGVRLSS-GQDLYSHKLILDPSFTIPSTPSLSPKDFSLQMLSHVDI 1193
Query: 295 VGKVARAICIMSHPIPDTNDSHSAQVILPQKQL 327
G VAR ICI I D + V+ P + L
Sbjct: 1194KGMVARGICITRSSIKP--DVSNCSVVYPPRSL 1286
>BE324242 weakly similar to GP|12322677|gb| Rab escort protein putative;
48705-51166 {Arabidopsis thaliana}, partial (10%)
Length = 478
Score = 31.2 bits (69), Expect = 0.84
Identities = 14/46 (30%), Positives = 28/46 (60%), Gaps = 1/46 (2%)
Frame = +1
Query: 67 SSREYNVDMI-PKFMMANGALVRVLIHTDVTKYLNFKAVDGSFVYN 111
+SR++N+D+ P+ + + + +L++ + NFK +D S VYN
Sbjct: 295 NSRKFNLDLAGPRALFSADKTIDLLLNQVLHSTYNFKGIDASLVYN 432
>AL375126 homologue to GP|3175990|emb| GDP dissociation inhibitor {Cicer
arietinum}, partial (6%)
Length = 404
Score = 30.8 bits (68), Expect = 1.1
Identities = 15/16 (93%), Positives = 16/16 (99%)
Frame = -3
Query: 427 GKVLDLSVDLSAASAA 442
GKVLDL+VDLSAASAA
Sbjct: 288 GKVLDLTVDLSAASAA 241
>TC85475 homologue to PIR|T06507|T06507 reversibly glycosylatable
polypeptide 1 - garden pea, partial (95%)
Length = 1726
Score = 28.5 bits (62), Expect = 5.5
Identities = 53/212 (25%), Positives = 76/212 (35%), Gaps = 36/212 (16%)
Frame = +1
Query: 109 VYNKGKIYKV--PATDVEALKSPLMGLFEKRRARKFFIYVQD---YEANDPKSHEGLDLN 163
+YN+ I ++ P + K F ++K +IY D + A DP HE L
Sbjct: 613 LYNRNDINRILGPKASCISFKDSACRCFGYMVSKKKYIYTIDDDCFVAKDPTGHEINALE 792
Query: 164 QVTARQLISK---------YGLEDDTVDFI---------------GHALALHLDD----S 195
Q + L+S Y D DF+ H L L++ D +
Sbjct: 793 Q-HIKNLLSPSTPFFFNTLYDPYRDGADFVRGYPFSLREGVPTAVSHGLWLNIPDYDAPT 969
Query: 196 YLDKPAKD---FVDRVKTYAESLARFQGGSPYIYPLYGLGELPQAFARLSAVYGGTYMLN 252
L KP + FVD V T + ++P+ G+ AF R L
Sbjct: 970 QLVKPHERNTRFVDAVMTIPKGT---------LFPMCGMN---LAFDR---------ELI 1086
Query: 253 KPECKVEFDGDGKAIGVTSDGETAKCKKVVCD 284
P GDG+ IG D C KV+CD
Sbjct: 1087GPAMYFGLMGDGQPIGRYDDMWAGWCIKVICD 1182
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.138 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,185,944
Number of Sequences: 36976
Number of extensions: 159847
Number of successful extensions: 617
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 612
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 615
length of query: 445
length of database: 9,014,727
effective HSP length: 99
effective length of query: 346
effective length of database: 5,354,103
effective search space: 1852519638
effective search space used: 1852519638
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144517.16


