

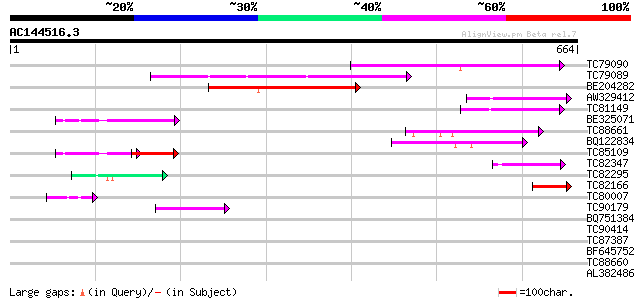
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144516.3 + phase: 0
(664 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC79090 similar to PIR|T46111|T46111 probable transposase - Arab... 179 4e-45
TC79089 weakly similar to PIR|T46111|T46111 probable transposase... 169 4e-42
BE204282 weakly similar to PIR|C85069|C850 hypothetical protein ... 164 8e-41
AW329412 weakly similar to GP|11994591|db Ac transposase-like pr... 88 9e-18
TC81149 weakly similar to GP|11994591|dbj|BAB02646. Ac transposa... 79 4e-15
BE325071 similar to GP|3928077|gb|A unknown protein {Arabidopsis... 79 7e-15
TC88661 weakly similar to GP|22507085|gb|AAM97760.1 putative tra... 72 7e-13
BQ122834 weakly similar to GP|20521440|dbj hypothetical protein~... 70 3e-12
TC85109 56 5e-08
TC82347 52 1e-06
TC82295 similar to PIR|A96722|A96722 unknown protein T17F3.2 [im... 49 8e-06
TC82166 similar to GP|10140707|gb|AAG13541.1 putative Tam3-trans... 44 2e-04
TC80007 44 3e-04
TC90179 weakly similar to GP|9719456|gb|AAF97810.1| transposase ... 44 3e-04
BQ751384 39 0.006
TC90414 37 0.032
TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I... 35 0.093
BF645752 homologue to GP|22713420|gb| Unknown (protein for MGC:3... 35 0.12
TC88660 PIR|T48044|T48044 hypothetical protein T12C14.220 - Arab... 28 0.37
AL382486 32 0.79
>TC79090 similar to PIR|T46111|T46111 probable transposase - Arabidopsis
thaliana, partial (28%)
Length = 1068
Score = 179 bits (453), Expect = 4e-45
Identities = 88/258 (34%), Positives = 150/258 (58%), Gaps = 8/258 (3%)
Frame = +2
Query: 400 WRRAEIMCEILKPFFTITNLISGSSYPTSNLYFGEIWKIECLIRSYLTSEDLLIQKMAEN 459
W+ EI+C LKP + N+++ ++YPT+ +F E+WK+ + + +ED I + +
Sbjct: 56 WKLVEILCTYLKPLYDAANILTTTTYPTAITFFHEVWKLHLDLARAVKNEDPFISDLTKP 235
Query: 460 MKVKFDKYWSDYNVVLAVGAVLDPTKKFNFLKFAYEKLDPLTSEEKLKKVKMTLGKLFSE 519
M K DKYW D ++ L + V+DP K ++F++ K+ + +K V + +LF E
Sbjct: 236 MYEKIDKYWRDCSLALVIAVVMDPRFKMKLVEFSFTKIYGEDAHAYVKIVDDGIHELFHE 415
Query: 520 YIKNGIP--------SNLSSSQVQPSYGGGTRITSSSYDEFEEYESQSSNNTGKSELDTY 571
Y +P N SS GGT ++ + +F+ Y ++S + KSELD Y
Sbjct: 416 YATLPLPLTPAYAEEGNAGSSMKMEGSSGGTLLSDNGLTDFDAYIMETSTHQTKSELDQY 595
Query: 572 LDELRMPLSQEFDVLAFWKERSRRSPNLARMACDILSIPITTVASESAFSIGARVVNRYR 631
L+E +P +FDVL +WK + P L++MA DILSIP++TV S+S F ++ +++YR
Sbjct: 596 LEESLLPRVPDFDVLGWWKLNKLKYPTLSKMARDILSIPVSTVPSDSIFDKKSKEMDQYR 775
Query: 632 SSMKDDSVQALLCARSWL 649
SS++ ++V+AL+CA+ W+
Sbjct: 776 SSLRPETVEALVCAKDWM 829
>TC79089 weakly similar to PIR|T46111|T46111 probable transposase -
Arabidopsis thaliana, partial (36%)
Length = 942
Score = 169 bits (427), Expect = 4e-42
Identities = 98/307 (31%), Positives = 161/307 (51%), Gaps = 1/307 (0%)
Frame = +3
Query: 165 VSRNTIAKEILNVYRDEKQKLKSQLAQIRGRVCLTSDCWTACSNEGYISLTAHYVNVNWK 224
V+ NTI + + Y EKQ L ++ GR LT D WT+ + GY+ +T H+V+ +WK
Sbjct: 21 VTFNTIQGDCVATYLTEKQNLSKYFDELPGRFGLTLDMWTSSQSVGYVFITGHFVDSDWK 200
Query: 225 LESKILAFAHMEP-PHSGRDLALKVLEMLDDWGIEKKIFSITLDNASANNSMANFLKEHL 283
L+ +IL MEP P S L+ V + DW +E ++F+IT + ++ N L+ L
Sbjct: 201 LQKRILNVV-MEPCPDSDSALSHAVSACISDWNLEGRLFTITCNQPLTKVALEN-LRPLL 374
Query: 284 SLSNSLLLDGEFFHIRCSAHILNLIVQDGLKVVSDALHKIRQSVAYVRVTEGRTLLFSEC 343
S+ N L+ +G+ C A L+ + D L ++KIR+SV YV+ +E F +
Sbjct: 375 SVKNPLIFNGQLLIGHCIARTLSNVAYDLLSSAQGIVNKIRESVKYVKTSESHEDKFLDL 554
Query: 344 VRIVGDIDTTIGLRLDCVTRWNSTYIMLQSALVYRRAFYSLSLRDSNFKCCPTSEEWRRA 403
+ + + L +D T+WN+TY ML +A + F L D ++K P+ +W+
Sbjct: 555 KEHL-QVPSEKSLFIDDQTKWNTTYQMLVAASELKEVFSCLDTSDPDYKGAPSIPDWKLV 731
Query: 404 EIMCEILKPFFTITNLISGSSYPTSNLYFGEIWKIECLIRSYLTSEDLLIQKMAENMKVK 463
EI+C LKP + N++ ++YPT+ +F E+ K+ + +ED I + + M K
Sbjct: 732 EILCTYLKPLYDAANILMTTTYPTAITFFHEVXKLHLDLSRAXKNEDPFISDLTKPMYXK 911
Query: 464 FDKYWSD 470
DKY D
Sbjct: 912 IDKYXRD 932
>BE204282 weakly similar to PIR|C85069|C850 hypothetical protein AT4g05510
[imported] - Arabidopsis thaliana, partial (16%)
Length = 563
Score = 164 bits (416), Expect = 8e-41
Identities = 86/185 (46%), Positives = 117/185 (62%), Gaps = 8/185 (4%)
Frame = +3
Query: 234 HMEPPHSGRDLALKVLEMLDDWGIEKKIFSITLDNASANNSMANFLKEHLSLSNSLL--- 290
H+EP H +L+ KV+ L +WGIEK IFSIT+DNASAN+ + LK+ L +SLL
Sbjct: 3 HLEPSHDNFELSRKVIGYLQEWGIEKNIFSITMDNASANDGNSQNLKDQLCSLDSLLRTF 182
Query: 291 -----LDGEFFHIRCSAHILNLIVQDGLKVVSDALHKIRQSVAYVRVTEGRTLLFSECVR 345
H +C AH+L+L+VQ+ LKVVSD L KIR S+ +V V+ R F +CV
Sbjct: 183 ITSATAPSLEKHFKCCAHVLDLMVQESLKVVSDVLDKIRNSLKFVSVSNSRLKQFCQCVE 362
Query: 346 IVGDIDTTIGLRLDCVTRWNSTYIMLQSALVYRRAFYSLSLRDSNFKCCPTSEEWRRAEI 405
VG D++ L LD +W STY+ML+SA+ YR AF + L+D+ + CP+SEEW R E
Sbjct: 363 EVGGGDSSDSLHLDVSGKWVSTYLMLKSAIKYRSAFEHMCLKDTTYNHCPSSEEWERGEK 542
Query: 406 MCEIL 410
+CE L
Sbjct: 543 ICEFL 557
>AW329412 weakly similar to GP|11994591|db Ac transposase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 569
Score = 88.2 bits (217), Expect = 9e-18
Identities = 48/123 (39%), Positives = 73/123 (59%), Gaps = 1/123 (0%)
Frame = +3
Query: 536 PSYGGGTRITSSSYDEFEEYESQSSNNTGKSELDTYLDELRMPLSQEFDVLAFWKERSRR 595
PS G R +D F SQS + T S+LD YL+E P + +F++L +WK + R
Sbjct: 33 PSTSSGARDRLKGFDRFLHETSQSQSMT--SDLDKYLEEPIFPRNSDFNILNWWKVHTPR 206
Query: 596 SPNLARMACDILSIPITTVASESAFSIGARVVNRYRSSMKDDSVQALLCARSWLHG-FEG 654
P L+ MA D+L P++T+A E AF+ G R+++ RSS+ D+ +AL+C WL EG
Sbjct: 207 YPILSMMARDVLGTPMSTLAPELAFNTGGRLLDSSRSSLNPDTREALICTHDWLRNESEG 386
Query: 655 ISS 657
++S
Sbjct: 387 LNS 395
>TC81149 weakly similar to GP|11994591|dbj|BAB02646. Ac transposase-like
protein {Arabidopsis thaliana}, partial (11%)
Length = 806
Score = 79.3 bits (194), Expect = 4e-15
Identities = 46/122 (37%), Positives = 71/122 (57%)
Frame = +1
Query: 528 NLSSSQVQPSYGGGTRITSSSYDEFEEYESQSSNNTGKSELDTYLDELRMPLSQEFDVLA 587
N SSS P +R +D+F SQ KS+LD YL+E P + +F++L
Sbjct: 154 NGSSSLSLPWSAKDSRDRLMGFDKFLHETSQGEG--AKSDLDKYLEEPLFPRNVDFNILN 327
Query: 588 FWKERSRRSPNLARMACDILSIPITTVASESAFSIGARVVNRYRSSMKDDSVQALLCARS 647
+WK + R P L+ MA ++L IP++ VA E AF+ RV++R SS+ +VQAL+C++
Sbjct: 328 WWKVHTPRYPVLSMMARNVLGIPMSKVAPELAFNYSGRVLDRDWSSLNPATVQALVCSQD 507
Query: 648 WL 649
W+
Sbjct: 508 WI 513
>BE325071 similar to GP|3928077|gb|A unknown protein {Arabidopsis thaliana},
partial (5%)
Length = 616
Score = 78.6 bits (192), Expect = 7e-15
Identities = 48/148 (32%), Positives = 73/148 (48%), Gaps = 3/148 (2%)
Frame = +2
Query: 54 DVWLYLVKVGIVDGVEKCRCKACHKLLTCESGSGTSHLKR--HVRSCSKTIKNHDVGEMM 111
+VW Y VK+ DG E C CK C K +G SHL + H+ C +K+
Sbjct: 203 NVWNYFVKIE-ADGKELCECKTCGKQFI-SGRTGISHLNQVQHITKCPLIMKSR------ 358
Query: 112 IDVEGKLRKKKFDPMANREFLARIMITHGAPFNMVEWKVFREYQKFLN-DDCVFVSRNTI 170
+ + K D R+ + ++ I PF EW FR + KF + ++ +SR+ +
Sbjct: 359 --IAKHFKLNKIDHRMVRDSVTQMTIKSYIPFQFAEWDEFRAFTKFASFNEARSLSRDNV 532
Query: 171 AKEILNVYRDEKQKLKSQLAQIRGRVCL 198
+++ VY EK KLK QLA ++G VCL
Sbjct: 533 VADVMKVYLLEKDKLKKQLAAVKGGVCL 616
>TC88661 weakly similar to GP|22507085|gb|AAM97760.1 putative transposase
{Oryza sativa (japonica cultivar-group)}, partial (4%)
Length = 521
Score = 72.0 bits (175), Expect = 7e-13
Identities = 49/172 (28%), Positives = 86/172 (49%), Gaps = 10/172 (5%)
Frame = +1
Query: 464 FDKYWSDY---NVVLAVGAVLDPTKKFNFLKFAYEKLDPLTSE---EKLKKVKMTLGKLF 517
+DKYW + N L G + DP KF ++++++ L S+ E+ V L KL+
Sbjct: 1 YDKYWGNVVKMNQFLYFGVIFDPRYKFGYIEWSFNDLYGAGSDIAKERAGSVSDNLFKLY 180
Query: 518 ----SEYIKNGIPSNLSSSQVQPSYGGGTRITSSSYDEFEEYESQSSNNTGKSELDTYLD 573
SE+ PS ++S V+ +++ D ++++ +++L+ YL
Sbjct: 181 NLYKSEHESFVGPSGSNNSSVEQPAVPKIPSLNTTADAYKKHLKTKETIVPQNDLERYLS 360
Query: 574 ELRMPLSQEFDVLAFWKERSRRSPNLARMACDILSIPITTVASESAFSIGAR 625
+ FD+L +WK+ R P LA M D+L+ P+++VASESA S G +
Sbjct: 361 DPPENDDPSFDILTWWKKNCVRYPVLATMVRDVLATPVSSVASESAXSTGRK 516
>BQ122834 weakly similar to GP|20521440|dbj hypothetical protein~similar to
transposase {Oryza sativa (japonica cultivar-group)},
partial (10%)
Length = 553
Score = 70.1 bits (170), Expect = 3e-12
Identities = 48/171 (28%), Positives = 81/171 (47%), Gaps = 12/171 (7%)
Frame = +2
Query: 448 SEDLLIQKMAENMKVKFDKYWSDYNVVLAVGAVLDPTKKFNFLKFAYEKLDPLTSEEKLK 507
S+D + + + FD+YW D ++LAV +DP K ++ + + +E ++
Sbjct: 17 SQDPFLSSLIFPLLKNFDQYWRDSCLILAVAVAMDPRHKMKLVESTFTTIFGENAEPWIR 196
Query: 508 KVKMTLGKLFSEY--------IKNGIPSNLSSSQVQPSYG---GGTRITSSSYDEFEEYE 556
V+ L +LF +Y NG + +P G G + +FE
Sbjct: 197 IVEDGLHELFIDYNTEMLHFTATNGDDVDEIMLNTEPYEGPVDGSLFVDEGGLSDFEFNI 376
Query: 557 SQ-SSNNTGKSELDTYLDELRMPLSQEFDVLAFWKERSRRSPNLARMACDI 606
S +S KSE+D YL+E + S+EFD+L++W+E + P L+R A DI
Sbjct: 377 SDFTSMQQFKSEMDEYLEEPLLSESKEFDILSWWRENRSKYPTLSRAASDI 529
>TC85109
Length = 559
Score = 55.8 bits (133), Expect = 5e-08
Identities = 34/102 (33%), Positives = 50/102 (48%), Gaps = 1/102 (0%)
Frame = +3
Query: 54 DVWLYLVKVGIVDGVEKCRCKACHKLLTCESGSGTSHLKRHVRSCSKTIKNHDVGEMMID 113
+VW Y VKV DG E C C C ++ TC+ SG SHL H+ C +K+
Sbjct: 147 NVWNYFVKVH-KDGKEMCECMTCGRVFTCDGRSGNSHLNLHIPVCPLVMKSR-------- 299
Query: 114 VEGKLRKKKFDPMANREFLARIMITHGAPFNMVEWKV-FREY 154
+ + KK D RE + ++ I + PF++ V FRE+
Sbjct: 300 IAEHFKLKKIDHRMVRESITQMTIKNHLPFSVC*VGVKFREF 425
Score = 43.1 bits (100), Expect = 3e-04
Identities = 24/57 (42%), Positives = 35/57 (61%), Gaps = 2/57 (3%)
Frame = +1
Query: 143 FNMVEWK-VFREYQKFLN-DDCVFVSRNTIAKEILNVYRDEKQKLKSQLAQIRGRVC 197
F VEW + KF++ ++ F+SR +A +++ VY EK+KLK QLA I G VC
Sbjct: 388 FRSVEWG*NLGNFTKFVSFNEAQFLSRYGVANDVMKVYLLEKEKLKKQLAAIEGGVC 558
>TC82347
Length = 750
Score = 51.6 bits (122), Expect = 1e-06
Identities = 29/87 (33%), Positives = 44/87 (50%), Gaps = 1/87 (1%)
Frame = +1
Query: 566 SELDTYLDELRMPLSQEFD-VLAFWKERSRRSPNLARMACDILSIPITTVASESAFSIGA 624
SELD Y R ++Q D + +W + P L+R+A DI +IP E AFS+
Sbjct: 328 SELDQYY---RQTIAQPVDNPVQWWISHRKTFPTLSRLALDIFAIPAMATDCERAFSLAK 498
Query: 625 RVVNRYRSSMKDDSVQALLCARSWLHG 651
+ R SMK +++ C ++WL G
Sbjct: 499 LTLTSQRLSMKSQTLEEAQCLQNWLRG 579
>TC82295 similar to PIR|A96722|A96722 unknown protein T17F3.2 [imported] -
Arabidopsis thaliana, partial (17%)
Length = 698
Score = 48.5 bits (114), Expect = 8e-06
Identities = 40/152 (26%), Positives = 59/152 (38%), Gaps = 40/152 (26%)
Frame = +1
Query: 73 CKACHKLLTCESGSGTSHLKRHVRSCSKTIKNHDVGEMMID----VEGKLR--------- 119
C+ C K L+ S SGTSHL+ H+ C + NH + + + EG L
Sbjct: 217 CRHCKKKLSGSSTSGTSHLRNHLIRCQRR-SNHGLAQYITAREKRKEGSLAITNFSLDQD 393
Query: 120 ---------------------------KKKFDPMANREFLARIMITHGAPFNMVEWKVFR 152
FD +R LAR++I HG P MVE FR
Sbjct: 394 PNKDDTVSLVNIKFEQAQLKDESVNTGNSNFDQRRSRFDLARMIILHGYPLAMVEHVGFR 573
Query: 153 EYQKFLNDDCVFVSRNTIAKEILNVYRDEKQK 184
+ K L V+ N + + + +Y E++K
Sbjct: 574 AFVKNLQPLFELVTLNRVEADCIEIYDKERKK 669
>TC82166 similar to GP|10140707|gb|AAG13541.1 putative Tam3-transposase
{Oryza sativa}, partial (6%)
Length = 574
Score = 44.3 bits (103), Expect = 2e-04
Identities = 22/47 (46%), Positives = 32/47 (67%), Gaps = 1/47 (2%)
Frame = +1
Query: 613 TVASESAFSIGARVVNRYRSSMKDDSVQALLCARSWLH-GFEGISSI 658
TVASESAFS R V RS +K+D+++AL+C+++W +G S I
Sbjct: 7 TVASESAFSTSGRDVTPQRSRLKEDTLEALMCSQNWFRTEMQGYSKI 147
>TC80007
Length = 1599
Score = 43.5 bits (101), Expect = 3e-04
Identities = 26/60 (43%), Positives = 33/60 (54%)
Frame = +3
Query: 44 DKSKGKALTSDVWLYLVKVGIVDGVEKCRCKACHKLLTCESGSGTSHLKRHVRSCSKTIK 103
+ SK +TS VW + +V +G K CK C KLL + GTSHLKRH+ C K K
Sbjct: 159 NSSKRPKMTSKVWDEMERVQTTEG-NKVLCKYCGKLL--QDNCGTSHLKRHLVICPKRPK 329
>TC90179 weakly similar to GP|9719456|gb|AAF97810.1| transposase
{Cryphonectria parasitica}, partial (3%)
Length = 810
Score = 43.5 bits (101), Expect = 3e-04
Identities = 20/87 (22%), Positives = 45/87 (50%)
Frame = -1
Query: 171 AKEILNVYRDEKQKLKSQLAQIRGRVCLTSDCWTACSNEGYISLTAHYVNVNWKLESKIL 230
A+E+ +Y ++ +K ++ Q +V L+ D WT+ + I++ H+++ +L
Sbjct: 393 ARELHQLYEAKRVIVKEEMRQALTKVHLSFDLWTSPNRLAIIAVFGHFISPAGNDSRYLL 214
Query: 231 AFAHMEPPHSGRDLALKVLEMLDDWGI 257
A HSG ++A + +++ DW +
Sbjct: 213 ALRRQPGAHSGENIASTICQVIRDWDL 133
>BQ751384
Length = 455
Score = 38.9 bits (89), Expect = 0.006
Identities = 22/97 (22%), Positives = 50/97 (50%), Gaps = 3/97 (3%)
Frame = -1
Query: 132 LARIMITHGAPFNMVEWKVFREYQKFLNDDCVFVSRNTIAKEILNVYRDEKQKLKSQL-A 190
+ +++ + F++++ K F++ + N ++R+ + + Y + +L A
Sbjct: 293 ILNLIVENNLSFSILDSKSFKDLLNYYNKSNPILNRHKVKNLLEFTYSRFLSTINYELEA 114
Query: 191 QIR--GRVCLTSDCWTACSNEGYISLTAHYVNVNWKL 225
I+ G+ LT D WT+ S + Y+S+ Y+N ++KL
Sbjct: 113 NIKSLGKFSLTLDIWTSKSQDTYLSIIISYINSSFKL 3
>TC90414
Length = 621
Score = 36.6 bits (83), Expect = 0.032
Identities = 21/54 (38%), Positives = 29/54 (52%), Gaps = 2/54 (3%)
Frame = +1
Query: 66 DGVEKCRCKACHKLLTCESGSGTSHLKRHVRSCSKTIKNHDVGEMM--IDVEGK 117
DG K CK C K L E+ +GTSHLK H+ C+ K + ++ +V GK
Sbjct: 415 DGKFKAICKYCDKKLGGETTNGTSHLKDHLSICAARNKRSPMQALLKVSEVPGK 576
>TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I3.230 -
Arabidopsis thaliana, partial (14%)
Length = 1074
Score = 35.0 bits (79), Expect = 0.093
Identities = 36/121 (29%), Positives = 55/121 (44%), Gaps = 1/121 (0%)
Frame = +3
Query: 3 VDGEIESQAREIPLVPPIDQNAQEVSSDPKDKKRKGKAKAKDKSKGKALTSDVWLYLVKV 62
VDGE + + +++ D A EV D K+KK K K KDK G A + +
Sbjct: 318 VDGEEKVKTKKV------DDAAVEVEDDKKEKK---KKKKKDKENGAAASDE-------- 446
Query: 63 GIVDGVEKCRCKACHKLLTCESG-SGTSHLKRHVRSCSKTIKNHDVGEMMIDVEGKLRKK 121
+ VEK + K HK E G G+ +++ + K + +VG +D K +KK
Sbjct: 447 ---EKVEKEK-KKKHK----EKGEDGSPEVEKSDKKKKKHKETSEVGSPEVDKSEKKKKK 602
Query: 122 K 122
K
Sbjct: 603 K 605
>BF645752 homologue to GP|22713420|gb| Unknown (protein for MGC:33586) {Homo
sapiens}, partial (1%)
Length = 321
Score = 34.7 bits (78), Expect = 0.12
Identities = 19/54 (35%), Positives = 29/54 (53%)
Frame = +3
Query: 132 LARIMITHGAPFNMVEWKVFREYQKFLNDDCVFVSRNTIAKEILNVYRDEKQKL 185
+AR++I PF++VE F K L SR T+A++ ++ DEK KL
Sbjct: 153 IARLVILSKLPFSVVESDRFNRLCKLLQPLWNIPSRRTVARDCFRMFXDEKFKL 314
>TC88660 PIR|T48044|T48044 hypothetical protein T12C14.220 - Arabidopsis
thaliana, partial (2%)
Length = 865
Score = 27.7 bits (60), Expect(2) = 0.37
Identities = 9/20 (45%), Positives = 15/20 (75%)
Frame = +3
Query: 630 YRSSMKDDSVQALLCARSWL 649
YRSS+ + +AL+C ++WL
Sbjct: 54 YRSSLSPEMAEALICTQNWL 113
Score = 23.9 bits (50), Expect(2) = 0.37
Identities = 10/15 (66%), Positives = 13/15 (86%)
Frame = +1
Query: 614 VASESAFSIGARVVN 628
VASESAFS G R+++
Sbjct: 7 VASESAFSTGGRILD 51
>AL382486
Length = 492
Score = 32.0 bits (71), Expect = 0.79
Identities = 16/55 (29%), Positives = 32/55 (58%)
Frame = +3
Query: 588 FWKERSRRSPNLARMACDILSIPITTVASESAFSIGARVVNRYRSSMKDDSVQAL 642
FW + P L+ +A D + +PI++ E +FSI +++ R ++ +S++AL
Sbjct: 255 FWLGMVNQLPLLSNIALDYIWLPISSCTVERSFSIYNTILDDDRQNLSLESLKAL 419
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,618,834
Number of Sequences: 36976
Number of extensions: 311585
Number of successful extensions: 1694
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 1670
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1687
length of query: 664
length of database: 9,014,727
effective HSP length: 102
effective length of query: 562
effective length of database: 5,243,175
effective search space: 2946664350
effective search space used: 2946664350
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144516.3


