

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
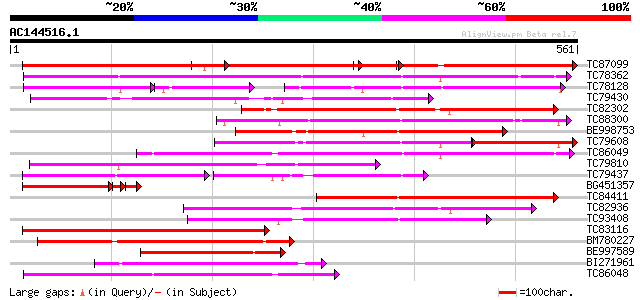
Query= AC144516.1 + phase: 0 /pseudo
(561 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87099 similar to PIR|G86449|G86449 hypothetical protein AAF813... 361 e-134
TC78362 similar to GP|15391731|emb|CAA93316. nitrite transporter... 318 3e-87
TC78128 weakly similar to GP|21928025|gb|AAM78041.1 At1g69870/T1... 185 3e-80
TC79430 similar to PIR|F86358|F86358 Similar to peptide transpor... 233 1e-61
TC82302 similar to PIR|F84663|F84663 probable nitrate transporte... 232 3e-61
TC88300 similar to GP|9581817|emb|CAC00544.1 putative low-affini... 229 2e-60
BE998753 similar to PIR|T45958|T45 oligopeptide transporter-like... 224 5e-59
TC79608 similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7 {... 157 2e-56
TC86049 weakly similar to GP|20466248|gb|AAM20441.1 putative tra... 214 5e-56
TC79810 similar to GP|11994403|dbj|BAB02362. nitrate transporter... 213 2e-55
TC79437 similar to PIR|G96720|G96720 nitrate transporter (NTL1) ... 117 2e-55
BG451357 similar to PIR|G86449|G86 hypothetical protein AAF81343... 180 3e-54
TC84411 similar to GP|4102839|gb|AAD01600.1| LeOPT1 {Lycopersico... 206 3e-53
TC82936 similar to PIR|G84829|G84829 probable PTR2 family peptid... 201 8e-52
TC93408 similar to GP|11933407|dbj|BAB19758. putative nitrate tr... 200 1e-51
TC83116 similar to GP|11933407|dbj|BAB19758. putative nitrate tr... 192 2e-49
BM780227 similar to PIR|E84798|E84 probable peptide/amino acid t... 177 1e-44
BE997589 weakly similar to PIR|G86449|G86 hypothetical protein A... 156 2e-38
BI271961 similar to PIR|T47573|T475 peptide transport-like prote... 147 1e-35
TC86048 weakly similar to PIR|H96561|H96561 probable peptide tra... 144 9e-35
>TC87099 similar to PIR|G86449|G86449 hypothetical protein AAF81343.1
[imported] - Arabidopsis thaliana, partial (86%)
Length = 2205
Score = 361 bits (926), Expect = e-100
Identities = 170/205 (82%), Positives = 191/205 (92%)
Frame = +2
Query: 13 VNQCLATLAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGR 72
+NQ LATLAFFG+GVNLV+FLTRV+GQ+NA AANNVSKWTGTVYIFSLVGAFLSDSYWGR
Sbjct: 227 LNQGLATLAFFGVGVNLVLFLTRVLGQDNAAAANNVSKWTGTVYIFSLVGAFLSDSYWGR 406
Query: 73 YKTCAVFQVIFVIGLMSLSLSSYLFLIKPKGCGNESIQCGKHSSWEMGMFYLSIYLIALG 132
YKTCA+FQ IFV GL+SLS+++YL L++PKGCGN ++CG+HSS EMGMFYLSIYLIALG
Sbjct: 407 YKTCAIFQGIFVTGLVSLSVTTYLALLRPKGCGNGKLECGEHSSLEMGMFYLSIYLIALG 586
Query: 133 NGGYQPNIATFGADQFDEEHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGMWAL 192
NGGYQPNIATFGADQFDE+HSKE ++KVAFFSYFYLALN G LFSNTIL YFEDEG+WAL
Sbjct: 587 NGGYQPNIATFGADQFDEDHSKESYSKVAFFSYFYLALNLGSLFSNTILGYFEDEGLWAL 766
Query: 193 GFWLSAGCAFIALILFLVGTPKYRH 217
GFW SAG AF+AL+LFLVGTPKYRH
Sbjct: 767 GFWASAGSAFLALVLFLVGTPKYRH 841
Score = 284 bits (727), Expect(2) = e-134
Identities = 143/179 (79%), Positives = 158/179 (87%)
Frame = +2
Query: 383 KKSDSKGLTELQRMGVGLVIAVIAMVSAGIVESYRLKHADQDCRHCKGSSTLSIFWQIPQ 442
K SKGLTELQRMG+GLVIA+IAMV+AGIVE YRLK+A Q +S+LSIFWQIPQ
Sbjct: 1340 KNQVSKGLTELQRMGIGLVIAIIAMVTAGIVECYRLKYAKQG-----DTSSLSIFWQIPQ 1504
Query: 443 YALIGASEVFMYVGQLEFFNAQTPDGLKSFGSALCMTSISLGNYVSSLLVSVVMKISTED 502
YALIGASEVFMYVGQLEFFNAQTPDGLKSFGSALCMTSISLGNYVSSL+VS+VMKISTED
Sbjct: 1505 YALIGASEVFMYVGQLEFFNAQTPDGLKSFGSALCMTSISLGNYVSSLIVSIVMKISTED 1684
Query: 503 HMPGWIPVNLNKGHLDRFYFLLAALTSMDLIAYIACAKWYKSIQLGEKRKENDEPGSFK 561
HMPGWIP NLN+GHLDRF+FLLA LTS+DLIAYIACAKW+++IQ+ K NDEP S K
Sbjct: 1685 HMPGWIPGNLNRGHLDRFFFLLAVLTSLDLIAYIACAKWFQNIQMACKYDNNDEPSSCK 1861
Score = 213 bits (542), Expect(2) = e-134
Identities = 112/179 (62%), Positives = 126/179 (69%), Gaps = 9/179 (5%)
Frame = +3
Query: 181 LVYFEDEGMWA---------LGFWLSAGCAFIALILFLVGTPKYRHFKPSGNPLARFCQV 231
LV+F WA LGF L G F L FL+ FKP GNPL+RFCQV
Sbjct: 705 LVHFSQTLFWAILKMKDYGLLGFGLLLGLPFWLLYFFLLAPQNIDTFKPCGNPLSRFCQV 884
Query: 232 LVAASRKVKVEMPSNGDDLYNMDTKESSTDVNRKILHTHGFKFLDRAAIISSRDHDNQKG 291
AASRK+ V+M SNGDDLY +D KESS + NRKILHTHGFKFLDRAA I+SRD + QKG
Sbjct: 885 FFAASRKLGVQMTSNGDDLYVIDEKESSNNSNRKILHTHGFKFLDRAAYITSRDLEVQKG 1064
Query: 292 TCSNPWRLCPVSQVEEVKCILRLLPIWLCTIIYSVVFTQMASLFVEQGDAMKTTVYNFR 350
NPW LCP++QVEEVKCILRLLPIWLCTIIYSVVFTQMASLFVEQG + + F+
Sbjct: 1065GQHNPWXLCPITQVEEVKCILRLLPIWLCTIIYSVVFTQMASLFVEQGACNENHNFQFQ 1241
Score = 80.5 bits (197), Expect = 2e-15
Identities = 36/49 (73%), Positives = 44/49 (89%)
Frame = +1
Query: 341 AMKTTVYNFRVPPASMSCFDIISVAVFIFFYRRILDPFVGKLKKSDSKG 389
AMKTT+ +F++PPASMS FDI+SVA+FIFFYRR+LDP VGKLKKS +G
Sbjct: 1213 AMKTTISSFKIPPASMSSFDILSVAIFIFFYRRVLDPLVGKLKKSSFQG 1359
>TC78362 similar to GP|15391731|emb|CAA93316. nitrite transporter {Cucumis
sativus}, partial (88%)
Length = 1871
Score = 318 bits (815), Expect = 3e-87
Identities = 178/547 (32%), Positives = 310/547 (56%), Gaps = 4/547 (0%)
Frame = +3
Query: 14 NQCLATLAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGRY 73
N+ A G N++ +LT+ + A+N +S ++G + L+GAF++DS+ GR+
Sbjct: 120 NEVCDRFASAGFHANMITYLTQQLNMPLVSASNTLSNFSGLSSLTPLLGAFIADSFAGRF 299
Query: 74 KTCAVFQVIFVIGLMSLSLSSYLFLIKPKGCGNESIQCGKHSSWEMGMFYLSIYLIALGN 133
T +I+ +GL++++ S+ + +P C + + C + S ++ + YL+++L +LG+
Sbjct: 300 WTIVFATLIYELGLITITTSAIVPHFRPPPCPTQ-VNCQEAKSSQLWILYLALFLTSLGS 476
Query: 134 GGYQPNIATFGADQFDEEHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGMWALG 193
GG + + F DQFD K F++++ + F L + TI+VY +D W G
Sbjct: 477 GGIRSCVVPFSGDQFDMTKKGVESRKWNLFNWYFFCMGFASLSALTIVVYIQDHMGWGWG 656
Query: 194 FWLSAGCAFIALILFLVGTPKYRHFKPSGNPLARFCQVLVAASRKVKVEMPSNGDDLYNM 253
+ FIA+I FL+G+ Y+ KPSG+P R QV+VAA RK +P++ LY
Sbjct: 657 LGIPTIAMFIAIIAFLLGSRLYKTLKPSGSPFLRLAQVIVAAFRKRNDALPNDPKLLYQN 836
Query: 254 DTKESSTDVNRKILHTHGFKFLDRAAIISSRDHDNQKGTCSNPWRLCPVSQVEEVKCILR 313
+SS + ++ HT +K+LD+AAI++ + N N W L V +VEE+KC++R
Sbjct: 837 LELDSSISLEGRLSHTDQYKWLDKAAIVTDEEAKN-LNKLPNLWNLATVHRVEELKCLVR 1013
Query: 314 LLPIWLCTIIYSVVFTQMASLFVEQGDAM-KTTVYNFRVPPASMSCFDIISVAVFIFFYR 372
+LPIW I+ + S + Q M + + F + PASM+ F ++++ + Y
Sbjct: 1014MLPIWASGILLITASSSQHSFVIVQARTMDRHLSHTFEISPASMAIFSVLTMMTGVILYE 1193
Query: 373 RILDPFVGKLKKSDSKGLTELQRMGVGLVIAVIAMVSAGIVESYRLKHADQD--CRHCKG 430
R+ PF+ + K+ + G+T LQRMG+G VI +IA + + ++E R K A + K
Sbjct: 1194RLFVPFIRRFTKNPA-GITCLQRMGIGFVINIIATIVSALLEIKRKKVASKYHLLDSPKA 1370
Query: 431 SSTLSIFWQIPQYALIGASEVFMYVGQLEFFNAQTPDGLKSFGSALCMTSISLGNYVSSL 490
+S+FW +PQY L G +EVFM VG LEF Q+P+ ++S +AL +I++G+++ +L
Sbjct: 1371IIPISVFWLVPQYFLHGVAEVFMNVGHLEFLYDQSPESMRSSATALYCIAIAIGHFIGTL 1550
Query: 491 LVSVVMKISTEDHMPGWIP-VNLNKGHLDRFYFLLAALTSMDLIAYIACAKWYKSIQLGE 549
LV++V K + ++ W+P NLN+G L+ +YFL+ + ++ I Y+ CA W + + E
Sbjct: 1551LVTLVHKYTGKER--NWLPDRNLNRGRLEYYYFLVCGIQVINFIYYVICA-WIYNYKPLE 1721
Query: 550 KRKENDE 556
+ EN++
Sbjct: 1722EINENNQ 1742
>TC78128 weakly similar to GP|21928025|gb|AAM78041.1 At1g69870/T17F3_10
{Arabidopsis thaliana}, partial (77%)
Length = 2022
Score = 185 bits (470), Expect(3) = 3e-80
Identities = 104/284 (36%), Positives = 165/284 (57%), Gaps = 6/284 (2%)
Frame = +2
Query: 273 KFLDRAAIISSRDHDNQKGTCSNPWRLCPVSQVEEVKCILRLLPIWLCTIIYSVVFTQMA 332
+ LD+AA++ D N GT N W L + QVEEVKC+ R LPIW I+ Q
Sbjct: 1058 RVLDKAALVMEGDL-NPDGTIVNQWNLVSIQQVEEVKCLARTLPIWAAGILGFTAMAQQG 1234
Query: 333 SLFVEQGDAMKTTVY---NFRVPPASMSCFDIISVAVFIFFYRRILDPFVGKLKKSDSKG 389
+ V Q AMK + F++P S+ +I + +++ FY RI P + K+ K++ G
Sbjct: 1235 TFIVSQ--AMKMDRHLGPKFQIPAGSLGVISLIVIGLWVPFYDRICVPSLRKITKNEG-G 1405
Query: 390 LTELQRMGVGLVIAVIAMVSAGIVESYRLKHADQDCRHCKGSSTLSIFWQIPQYALIGAS 449
+T LQR+G+G+V ++I+M+ AG+VE R A+ + +G + +S+ W PQ L+G
Sbjct: 1406 ITLLQRIGIGMVFSIISMIVAGLVEKVRRDVANSNPTP-QGIAPMSVMWLFPQLVLMGFC 1582
Query: 450 EVFMYVGQLEFFNAQTPDGLKSFGSALCMTSISLGNYVSSLLVSVVMKISTEDHMPGWIP 509
E F +G +EFFN Q PD ++S +AL S +L NYVSS+LV V ++ + P W+
Sbjct: 1583 EAFNIIGLIEFFNRQFPDHMRSIANALFSCSFALANYVSSILVITVHSVTKTHNHPDWLT 1762
Query: 510 VNLNKGHLDRFYFLLAALTSMDLIAYIACAKWYK---SIQLGEK 550
N+N+G LD FY+LLA + ++L+ ++ ++ Y S+ + EK
Sbjct: 1763 NNINEGRLDYFYYLLAGVGVLNLVYFLYVSQRYHYKGSVDIQEK 1894
Score = 77.4 bits (189), Expect(3) = 3e-80
Identities = 40/134 (29%), Positives = 74/134 (54%), Gaps = 3/134 (2%)
Frame = +2
Query: 14 NQCLATLAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGRY 73
N+ LA FG+ N +V+LTR A+N ++ W G L+GAF+SD+Y GR+
Sbjct: 266 NESFERLAAFGLLANFMVYLTREFHLEQVHASNILNIWGGISNFAPLLGAFISDTYTGRF 445
Query: 74 KTCAVFQVIFVIGLMSLSLSSYLFLIKPKGCGNES---IQCGKHSSWEMGMFYLSIYLIA 130
KT A ++G+ +++L+++L ++P C + QC +S ++G ++ + ++
Sbjct: 446 KTIAFASFFSLLGMTAVTLTAWLPKLQPPSCTPQQQALNQCVTANSSQVGFLFMGLIFLS 625
Query: 131 LGNGGYQPNIATFG 144
+G+ G +P FG
Sbjct: 626 IGSSGIRPCSIPFG 667
Score = 75.9 bits (185), Expect(3) = 3e-80
Identities = 40/101 (39%), Positives = 58/101 (56%), Gaps = 2/101 (1%)
Frame = +3
Query: 144 GADQFDE--EHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGMWALGFWLSAGCA 201
G DQFD E K+G N +FF+++Y + LF+ T++VY +D W GF + C
Sbjct: 666 GVDQFDPTTEKGKKGIN--SFFNWYYTSFTVVLLFTQTVIVYIQDSISWKFGFAIPTLCM 839
Query: 202 FIALILFLVGTPKYRHFKPSGNPLARFCQVLVAASRKVKVE 242
++ILF +GT Y H KP G+ + QVLVA+ +K K E
Sbjct: 840 LXSIILFFIGTKIYVHVKPEGSIFSSIAQVLVASFKKRKSE 962
>TC79430 similar to PIR|F86358|F86358 Similar to peptide transporter
[imported] - Arabidopsis thaliana, partial (55%)
Length = 1303
Score = 233 bits (594), Expect = 1e-61
Identities = 144/405 (35%), Positives = 222/405 (54%), Gaps = 6/405 (1%)
Frame = +2
Query: 21 AFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGRYKTCAVFQ 80
A++GI NL+ +LT +GQ+ A AA NV+ W+GT + L+GAFL+DS+ GRY+T +
Sbjct: 188 AYYGINSNLINYLTGPLGQSTATAAENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIAS 367
Query: 81 VIFVIGLMSLSLSSYLFLIKPKGCGNESIQCGKHSSWEMGMFYLSIYLIALGNGGYQPNI 140
+++++ L L+LS+ L P ++I +F+ S+YL+A GG++P +
Sbjct: 368 LVYILALSLLTLSATL----PSDGDAQAI-----------LFFFSLYLVAFAQGGHKPCV 502
Query: 141 ATFGADQFDEEHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGMWALGFWLSAGC 200
FGADQFD H +E ++ +FF+++Y G L + +IL Y +D W LGF +
Sbjct: 503 QAFGADQFDINHPQERRSRSSFFNWWYFTFTAGLLVTVSILNYVQDNVGWVLGFGIPWIA 682
Query: 201 AFIALILFLVGTPKYRHFKPSG---NPLARFCQVLVAASRKVKVEMPSNGDDLYNMDTKE 257
IAL LF +GT YR P P +R +V + A R N T E
Sbjct: 683 MIIALSLFSLGTWTYRFSIPGNQQRGPFSRIGRVFITALR--------------NFRTTE 820
Query: 258 SSTDVNRKILH--THGFKFLDRAAIISSRDHDNQKGTCSNPWRLCPVSQVEEVKCILRLL 315
+ +LH + F FL++A I S +N K +C VS+VEE K ILRL+
Sbjct: 821 EE-EPRPSLLHQPSQQFSFLNKALIASDGSKENGK--------VCSVSEVEEAKAILRLV 973
Query: 316 PIWLCTIIYSVVFTQMASLFVEQGDAMKTTVY-NFRVPPASMSCFDIISVAVFIFFYRRI 374
PIW ++I+++VF+Q ++ F +QG + + F VPPAS+ F +S+ +FI Y RI
Sbjct: 974 PIWATSLIFAIVFSQSSTFFTKQGVTLDRKILPGFYVPPASLQSFISLSIVLFIPVYDRI 1153
Query: 375 LDPFVGKLKKSDSKGLTELQRMGVGLVIAVIAMVSAGIVESYRLK 419
+ P + + G+T LQR+G G++ +VI+MV A VE R K
Sbjct: 1154IVP-IARTFTGKPSGITMLQRIGAGILFSVISMVIAAFVEMKRSK 1285
>TC82302 similar to PIR|F84663|F84663 probable nitrate transporter
[imported] - Arabidopsis thaliana, partial (51%)
Length = 1133
Score = 232 bits (591), Expect = 3e-61
Identities = 127/318 (39%), Positives = 198/318 (61%), Gaps = 4/318 (1%)
Frame = +3
Query: 230 QVLVAASRKVKVEMPSNGDDLYNMDTKESSTDVNRKILHTHGFKFLDRAAIISSRDHDNQ 289
QV+VA+ +K K+E+P N LY DT E S +I + F+FL++AAI+ D D
Sbjct: 9 QVIVASIKKRKMELPYNVGSLYE-DTPEDS-----RIEQSEQFRFLEKAAIVVEGDFDKD 170
Query: 290 K-GTCSNPWRLCPVSQVEEVKCILRLLPIWLCTIIYSVVFTQMASLFVEQGDAMKTTVYN 348
G+ NPW+LC +++VEEVK ++RLLPIW TII+ + QM + VEQ M+ V N
Sbjct: 171 LYGSGPNPWKLCSLTRVEEVKMMVRLLPIWATTIIFWTTYAQMITFSVEQAATMERNVGN 350
Query: 349 FRVPPASMSCFDIISVAVFIFFYRRILDPFVGKLKKSDSKGLTELQRMGVGLVIAVIAMV 408
F++P S++ F ++++ + + RI+ P KL + G + LQR +GL+++ I M
Sbjct: 351 FQIPAGSLTVFFVVAILLTLAVNDRIIMPLWKKL--NGKPGFSNLQRNAIGLLLSTIGMA 524
Query: 409 SAGIVESYRLKHADQDCRHCKGSST---LSIFWQIPQYALIGASEVFMYVGQLEFFNAQT 465
+A ++E RL A + KG+ T +S+F +PQ+ L+G+ E F+Y GQL+FF Q+
Sbjct: 525 AASLIEVKRLSVA----KGVKGNQTTLPISVFLLVPQFFLVGSGEAFIYTGQLDFFITQS 692
Query: 466 PDGLKSFGSALCMTSISLGNYVSSLLVSVVMKISTEDHMPGWIPVNLNKGHLDRFYFLLA 525
P G+K+ + L +T++SLG +VSS LVSVV K++ GW+ ++NKG LD FY LL
Sbjct: 693 PKGMKTMSTGLFLTTLSLGFFVSSFLVSVVKKVTGTRDGQGWLADHINKGRLDLFYALLT 872
Query: 526 ALTSMDLIAYIACAKWYK 543
L+ ++ +A++ CA WYK
Sbjct: 873 ILSFINFVAFLVCAFWYK 926
>TC88300 similar to GP|9581817|emb|CAC00544.1 putative low-affinity nitrate
transporter {Nicotiana plumbaginifolia}, partial (56%)
Length = 1169
Score = 229 bits (584), Expect = 2e-60
Identities = 138/361 (38%), Positives = 216/361 (59%), Gaps = 9/361 (2%)
Frame = +1
Query: 205 LILFLVG--TPKYRHFKPSGNPLARFCQVLVAASRKVKVEMPSNGDDLYNMDTKESSTDV 262
L+LF+ + R K +G+PL + V VAA RK +E+P + L+N+D E
Sbjct: 1 LLLFVCSYWNKEVRFKKLAGSPLTQIAVVYVAAWRKRNMELPYDSSLLFNVDDIEDEMLR 180
Query: 263 NRK--ILHTHGFKFLDRAAIISSRDHDNQKGTCSNPWRLCPVSQVEEVKCILRLLPIWLC 320
+K + H+ F+FLD+AAI + N+ W L ++ VEEVK +LR+LPIW
Sbjct: 181 KKKQVLPHSKQFRFLDKAAIKDPKTDGNEINVVRK-WYLSTLTDVEEVKLVLRMLPIWAT 357
Query: 321 TIIYSVVFTQMASLFVEQGDAMKTTV-YNFRVPPASMSCFDIISVAVFIFFYRRILDPFV 379
TI++ V+ QM + V Q + + +F++PPAS++ F I S+ + I Y R++ P
Sbjct: 358 TIMFWTVYAQMTTFSVSQATTLNRHIGKSFQIPPASLTAFFIGSILLTIPIYDRVIVPIT 537
Query: 380 GKLKKSDSKGLTELQRMGVGLVIAVIAMVSAGIVESYRLKHAD-QDCRHCKGSST-LSIF 437
K+ K+ +GLT LQR+GVGLV ++ AMV+A + E R++ A + H S +S+F
Sbjct: 538 RKIFKNP-QGLTPLQRIGVGLVFSIFAMVAAALTELKRMRMAHLHNLTHNPNSEIPMSVF 714
Query: 438 WQIPQYALIGASEVFMYVGQLEFFNAQTPDGLKSFGSALCMTSISLGNYVSSLLVSVVMK 497
W IPQ+ +G+ E F Y+GQL+FF + P G+K+ + L ++++SLG + SSLLV++V K
Sbjct: 715 WLIPQFFFVGSGEAFTYIGQLDFFLRECPKGMKTMSTGLFLSTLSLGFFFSSLLVTLVHK 894
Query: 498 ISTEDHMPGWIPVNLNKGHLDRFYFLLAALTSMDLIAYIACAKW--YKSIQLGEKRKEND 555
+ T H P W+ NLN+ L FY+LLA L+ ++L+ Y+ CA W YK +L E+ E +
Sbjct: 895 V-TSQHKP-WLADNLNEAKLYNFYWLLALLSVLNLVIYLLCANWYVYKDKRLAEEGIELE 1068
Query: 556 E 556
E
Sbjct: 1069E 1071
>BE998753 similar to PIR|T45958|T45 oligopeptide transporter-like protein -
Arabidopsis thaliana, partial (44%)
Length = 809
Score = 224 bits (572), Expect = 5e-59
Identities = 123/271 (45%), Positives = 177/271 (64%), Gaps = 2/271 (0%)
Frame = +3
Query: 224 PLARFCQVLVAASRKVKVEMPSNGDDLYNMDTKESSTDVNRKILHTHGFKFLDRAAIISS 283
P + V+VA+ RK +V+ P++ LY + ES+ +RK+ HT+ +F D+AA+
Sbjct: 9 PPYSYRSVIVASIRKYRVDAPTDKSLLYEIADTESAIKGSRKLDHTNELRFFDKAAV--Q 182
Query: 284 RDHDNQKGTCSNPWRLCPVSQVEEVKCILRLLPIWLCTIIYSVVFTQMASLFVEQGDAMK 343
+ DN K + NPWRLC V+QVEE+K ILRLLP+W II++ V+ QM++LFV QG M
Sbjct: 183 GESDNLKESI-NPWRLCTVTQVEELKSILRLLPVWATGIIFATVYGQMSTLFVLQGQTMN 359
Query: 344 TTVYN--FRVPPASMSCFDIISVAVFIFFYRRILDPFVGKLKKSDSKGLTELQRMGVGLV 401
T V N F++PPAS+S FD ISV ++ Y RI+ P K GLT+LQRMGVGL
Sbjct: 360 THVGNSSFKIPPASLSIFDTISVIFWVPVYDRIIVPIARKFT-GHKNGLTQLQRMGVGLF 536
Query: 402 IAVIAMVSAGIVESYRLKHADQDCRHCKGSSTLSIFWQIPQYALIGASEVFMYVGQLEFF 461
I++ +MV+A +E RL+ ++ + ++IFWQ+PQY LIG +EVF ++GQLEFF
Sbjct: 537 ISIFSMVAAAFLELVRLRTVRRNNYYELEEIPMTIFWQVPQYFLIGCAEVFTFIGQLEFF 716
Query: 462 NAQTPDGLKSFGSALCMTSISLGNYVSSLLV 492
Q PD ++S SAL + +++ G Y+SSLLV
Sbjct: 717 YEQAPDAMRSLCSALSLLTVAFGQYLSSLLV 809
>TC79608 similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7 {Arabidopsis
thaliana}, partial (59%)
Length = 1489
Score = 157 bits (396), Expect(2) = 2e-56
Identities = 95/263 (36%), Positives = 143/263 (54%), Gaps = 3/263 (1%)
Frame = +2
Query: 203 IALILFLVGTPKYRHFKPSGNPLARFCQVLVAASRKVKVEMPSNGDDLYNMDTKESSTDV 262
I++I F+ G P YR+ P G+P R +V VAA RK K+ N LY D ++S +
Sbjct: 29 ISIIAFIGGYPLYRNLNPEGSPFTRLVKVGVAAFRKKKIPKVPNSTLLYQNDELDASITL 208
Query: 263 NRKILHTHGFKFLDRAAIISSRDHDNQKGTCSNPWRLCPVSQVEEVKCILRLLPIWLCTI 322
K+LH+ KFLD+AAI++ + DN K + WRL V +VEE+K I+R+ PIW I
Sbjct: 209 GGKLLHSDQLKFLDKAAIVT--EEDNTK--TPDLWRLSTVHRVEELKSIIRMGPIWASGI 376
Query: 323 IYSVVFTQMASLFVEQGDAM-KTTVYNFRVPPASMSCFDIISVAVFIFFYRRILDPFVGK 381
+ + Q + ++Q M + +F +P SMS F I+++ Y R+L +
Sbjct: 377 LLITAYAQQGTFSLQQAKTMNRHLTKSFEIPAGSMSVFTILTMLFTTALYDRVLIRVARR 556
Query: 382 LKKSDSKGLTELQRMGVGLVIAVIAMVSAGIVESYRLKHADQD--CRHCKGSSTLSIFWQ 439
D +G+T L RMG+G VI++ A AG VE R K A + H +S+FW
Sbjct: 557 FTGLD-RGITFLHRMGIGFVISLFATFVAGFVEMKRKKVAMEHGLIEHSSEIIPISVFWL 733
Query: 440 IPQYALIGASEVFMYVGQLEFFN 462
+PQY+L G +E FM +G + F+
Sbjct: 734 VPQYSLHGMAEAFMSIGAFKSFS 802
Score = 81.3 bits (199), Expect(2) = 2e-56
Identities = 45/106 (42%), Positives = 65/106 (60%), Gaps = 3/106 (2%)
Frame = +1
Query: 459 EFFNAQTPDGLKSFGSALCMTSISLGNYVSSLLVSVVMKISTEDHMPGWIP-VNLNKGHL 517
EFF Q P+ + S A TSISLGNY+S+LLV++V K + + W+P NLNKG L
Sbjct: 793 EFFYDQAPESMTSTAMAFFWTSISLGNYISTLLVTLVHKFTKGPNGTNWLPDNNLNKGKL 972
Query: 518 DRFYFLLAALTSMDLIAYIACAKW--YKSIQLGEKRKENDEPGSFK 561
+ FY+L+ L ++LI Y+ CAK YK IQ+ K + + E + +
Sbjct: 973 EYFYWLITLLQFINLIYYLICAKMYTYKQIQVHHKGENSSEDNNIE 1110
>TC86049 weakly similar to GP|20466248|gb|AAM20441.1 putative transport
protein {Arabidopsis thaliana}, partial (43%)
Length = 1836
Score = 214 bits (546), Expect = 5e-56
Identities = 128/439 (29%), Positives = 227/439 (51%), Gaps = 5/439 (1%)
Frame = +2
Query: 126 IYLIALGNGGYQPNIATFGADQFDEEHSKEGHNKVA-FFSYFYLALNFGQLFSNTILVYF 184
+ LIA+GNGG +++ FGA Q + + + +G+ + FFS++Y + + T +VY
Sbjct: 2 LILIAIGNGGITCSLS-FGAYQVNRKDNPDGYRVLEIFFSWYYAFTTIAVIIALTGIVYI 178
Query: 185 EDEGMWALGFWLSAGCAFIALILFLVGTPKYRHFKPSGNPLARFCQVLVAASRKVKVEMP 244
+D W +GF + A I+ +LF + +P Y K + F QV VAA + K+ +P
Sbjct: 179 QDHLGWKVGFGVPAILMLISTVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLP 358
Query: 245 SNGDDLYNMDTKESSTDVNRKILHTHGFKFLDRAAIISSRDHD-NQKGTCSNPWRLCPVS 303
+ K+S ++ T +FL++A +I + D G+ N W LC V
Sbjct: 359 PKTSPEFYHQQKDSEL-----VVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVD 523
Query: 304 QVEEVKCILRLLPIWLCTIIYSV-VFTQMASLFVEQGDAMKTTVYNFRVPPASMSCFDII 362
QVEE+K I++++P+W I S+ + L + D + NF VP S S I+
Sbjct: 524 QVEELKAIIKVIPLWSTAITMSINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIV 703
Query: 363 SVAVFIFFYRRILDPFVGKLKKSDSKGLTELQRMGVGLVIAVIAMVSAGIVESYRLKHAD 422
++ ++I Y R+L P K++ ++ +RMG+GL + +++A I E+ R K A
Sbjct: 704 AILIWIIIYDRVLIPLASKIRGKPVI-ISPKKRMGIGLFFNFLHLITAAIFETVRRKEAI 880
Query: 423 QD--CRHCKGSSTLSIFWQIPQYALIGASEVFMYVGQLEFFNAQTPDGLKSFGSALCMTS 480
++ G +S W PQ L G +E+F +GQ EF+ + P + S ++L +
Sbjct: 881 KEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLA 1060
Query: 481 ISLGNYVSSLLVSVVMKISTEDHMPGWIPVNLNKGHLDRFYFLLAALTSMDLIAYIACAK 540
+ +GN VSSL++S++ + GW+ N+NKGH D++Y+++ + +++L+ Y+ C+
Sbjct: 1061MGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCS- 1237
Query: 541 WYKSIQLGEKRKENDEPGS 559
W + E + E GS
Sbjct: 1238WAYGPTVDEVSNVSKENGS 1294
>TC79810 similar to GP|11994403|dbj|BAB02362. nitrate transporter
{Arabidopsis thaliana}, partial (60%)
Length = 1236
Score = 213 bits (541), Expect = 2e-55
Identities = 119/351 (33%), Positives = 197/351 (55%), Gaps = 3/351 (0%)
Frame = +3
Query: 20 LAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGRYKTCAVF 79
+ GI +NLV +L + ++A +A V+ + GT+ + L+G FL+D+ GRY T A+
Sbjct: 195 ICVMGISMNLVTYLVGDLHLHSASSATIVTNFMGTLNLLGLLGGFLADAKLGRYLTVAIS 374
Query: 80 QVIFVIGLMSLSLSSYLFLIKPKGCGN---ESIQCGKHSSWEMGMFYLSIYLIALGNGGY 136
I +G+ L+L++ + + P C + QC + S ++ + + ++Y IALG GG
Sbjct: 375 ATIATVGVCMLTLATTIPSMTPPPCSEVRRQHHQCIQASGKQLSLLFAALYTIALGGGGI 554
Query: 137 QPNIATFGADQFDEEHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGMWALGFWL 196
+ N++ FG+DQFD KE N + FF+ FY ++ G LFS +LVY +D G+ +
Sbjct: 555 KSNVSGFGSDQFDTNDPKEEKNMIFFFNRFYFFISIGSLFSVVVLVYVQDNIGRGWGYGI 734
Query: 197 SAGCAFIALILFLVGTPKYRHFKPSGNPLARFCQVLVAASRKVKVEMPSNGDDLYNMDTK 256
SAG +A+ + L GTP YR KP G+PL +VL A +K + +PS D
Sbjct: 735 SAGTMLVAVGILLCGTPLYRFKKPQGSPLTIIWRVLFLAWKKRTLPIPS--------DPT 890
Query: 257 ESSTDVNRKILHTHGFKFLDRAAIISSRDHDNQKGTCSNPWRLCPVSQVEEVKCILRLLP 316
+ + K+ +T F+ LD+AAI+ + ++ G NPW + ++QVEEVK +++LLP
Sbjct: 891 LLNGYLEAKVTYTDKFRSLDKAAILD--ETKSKDGNNENPWLVSTMTQVEEVKMVIKLLP 1064
Query: 317 IWLCTIIYSVVFTQMASLFVEQGDAMKTTVYNFRVPPASMSCFDIISVAVF 367
IW I++ V++QM + +EQ M V + +P S+S F I++ +F
Sbjct: 1065IWSTCILFWTVYSQMNTFTIEQATFMNRKVGSLEIPAGSLSAFLFITILLF 1217
>TC79437 similar to PIR|G96720|G96720 nitrate transporter (NTL1)
53025-56402 [imported] - Arabidopsis thaliana, partial
(61%)
Length = 1424
Score = 117 bits (294), Expect(2) = 2e-55
Identities = 72/228 (31%), Positives = 126/228 (54%), Gaps = 15/228 (6%)
Frame = +1
Query: 202 FIALILFLVGTPKYRHFKPSGNPLARFCQVLVAASRKVKVEMPSNGDDLYNMDTKES--S 259
F ++ +FL G+ YR+ PSG+PL +VL+AA+ ++ + NM + S S
Sbjct: 709 FGSIPVFLAGSTTYRNKVPSGSPLTTILKVLIAATLNSCCTNKNSCSAVVNMSSSPSNPS 888
Query: 260 TDVNRKILH------------THGFKFLDRAAIISSRDHDNQKGTCSNPWRLCPVSQVEE 307
T + LH + KFL++A I + H + + C + Q+E+
Sbjct: 889 TQAKEQELHQTLKPTTPSQIPSESLKFLNKA-ITNKPIHSSLQ---------CTIQQLED 1038
Query: 308 VKCILRLLPIWLCTIIYSVVFTQMASLFVEQGDAMKTTVY-NFRVPPASMSCFDIISVAV 366
VK + ++ PI+ CTI+ + Q+++ VEQ M TT++ +F+VPPAS+ F ++ + +
Sbjct: 1039VKLVFKIFPIFACTIMLNACLAQLSTFSVEQAATMNTTLFSSFKVPPASLPVFPVLFLMI 1218
Query: 367 FIFFYRRILDPFVGKLKKSDSKGLTELQRMGVGLVIAVIAMVSAGIVE 414
Y I+ P+ K+ KS++ G+T LQR+G+GL+++++AM A IVE
Sbjct: 1219LAPIYDHIIIPYARKVTKSEA-GITHLQRIGIGLILSIVAMAIAAIVE 1359
Score = 116 bits (291), Expect(2) = 2e-55
Identities = 62/185 (33%), Positives = 105/185 (56%)
Frame = +3
Query: 13 VNQCLATLAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGR 72
V + L LA+ NLV++L + + +++ANNV+ + GT ++ +L+G FLSD++
Sbjct: 144 VVEILENLAYLANASNLVLYLKEYMHMSPSKSANNVTNFMGTSFLLALLGGFLSDAFLTT 323
Query: 73 YKTCAVFQVIFVIGLMSLSLSSYLFLIKPKGCGNESIQCGKHSSWEMGMFYLSIYLIALG 132
Y + VI +IGL+ L++ +++ +KP C NES C + + + M + +YL+ALG
Sbjct: 324 YHVYLISAVIELIGLIILTIQAHVPSLKPTKC-NESTPCEEVNGGKAAMLFAGLYLVALG 500
Query: 133 NGGYQPNIATFGADQFDEEHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGMWAL 192
GG + ++ G +QFDE + FF+YF L+ G L + T +V+ ED W
Sbjct: 501 VGGIKGSLPVHGGEQFDEATPNGRKQRSTFFNYFVFCLSCGALIAVTFVVWVEDNKGWEW 680
Query: 193 GFWLS 197
GF +S
Sbjct: 681 GFAIS 695
>BG451357 similar to PIR|G86449|G86 hypothetical protein AAF81343.1
[imported] - Arabidopsis thaliana, partial (20%)
Length = 606
Score = 180 bits (456), Expect(3) = 3e-54
Identities = 89/90 (98%), Positives = 90/90 (99%)
Frame = +1
Query: 13 VNQCLATLAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGR 72
+NQCLATLAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGR
Sbjct: 250 LNQCLATLAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGR 429
Query: 73 YKTCAVFQVIFVIGLMSLSLSSYLFLIKPK 102
YKTCAVFQVIFVIGLMSLSLSSYLFLIKPK
Sbjct: 430 YKTCAVFQVIFVIGLMSLSLSSYLFLIKPK 519
Score = 37.0 bits (84), Expect(3) = 3e-54
Identities = 16/16 (100%), Positives = 16/16 (100%)
Frame = +3
Query: 115 SSWEMGMFYLSIYLIA 130
SSWEMGMFYLSIYLIA
Sbjct: 558 SSWEMGMFYLSIYLIA 605
Score = 34.7 bits (78), Expect(3) = 3e-54
Identities = 13/13 (100%), Positives = 13/13 (100%)
Frame = +2
Query: 102 KGCGNESIQCGKH 114
KGCGNESIQCGKH
Sbjct: 518 KGCGNESIQCGKH 556
>TC84411 similar to GP|4102839|gb|AAD01600.1| LeOPT1 {Lycopersicon
esculentum}, partial (38%)
Length = 804
Score = 206 bits (523), Expect = 3e-53
Identities = 103/241 (42%), Positives = 163/241 (66%), Gaps = 1/241 (0%)
Frame = +1
Query: 304 QVEEVKCILRLLPIWLCTIIYSVVFTQMASLFVEQGDAMKTTVYNFRVPPASMSCFDIIS 363
QVEE+K ++R+ PIW II+S V+ QM++LFVEQG M T++ +F++ PAS+S F++ S
Sbjct: 1 QVEELKILIRMFPIWATGIIFSSVYAQMSTLFVEQGTMMNTSIGSFKLSPASLSTFEVAS 180
Query: 364 VAVFIFFYRRILDPFVGKLKKSDSKGLTELQRMGVGLVIAVIAMVSAGIVESYRLKHADQ 423
V +++ Y +IL P V K +G++ QR+G+G I+ + M++A VE RL+ A +
Sbjct: 181 VVMWVPVYDKILVPIVKKFT-GKKRGISVFQRIGIGPFISGLCMLAAAAVEIKRLQLARE 357
Query: 424 -DCRHCKGSSTLSIFWQIPQYALIGASEVFMYVGQLEFFNAQTPDGLKSFGSALCMTSIS 482
D LS+ WQIPQY ++GA+E+F +VGQLEFF ++PD +++ AL + + S
Sbjct: 358 LDLVDKPVGVPLSVLWQIPQYLILGAAEIFTFVGQLEFFYEESPDAMRTICGALPLLNFS 537
Query: 483 LGNYVSSLLVSVVMKISTEDHMPGWIPVNLNKGHLDRFYFLLAALTSMDLIAYIACAKWY 542
LGNY+SS ++++V +T+ GWIP NLNKGHLD ++ LL+ L+ ++++ +I AK Y
Sbjct: 538 LGNYLSSFILTIVTYFTTKGGRLGWIPDNLNKGHLDYYFLLLSGLSLLNMLVFIVAAKIY 717
Query: 543 K 543
K
Sbjct: 718 K 720
>TC82936 similar to PIR|G84829|G84829 probable PTR2 family peptide
transporter [imported] - Arabidopsis thaliana, partial
(53%)
Length = 1112
Score = 201 bits (510), Expect = 8e-52
Identities = 120/355 (33%), Positives = 197/355 (54%), Gaps = 6/355 (1%)
Frame = +3
Query: 173 GQLFSNTILVYFEDEGMWALGFWLSAGCAFIALILFLVGTPKYRH-FKPSGNPLARFCQV 231
G L + LVY ++ W LG+ + ++L++F +GTP YRH + S +P +V
Sbjct: 78 GALIATLGLVYIQENLGWGLGYGIPTAGLILSLVIFYIGTPIYRHKVRTSKSPAKDIIRV 257
Query: 232 LVAASRKVKVEMPSNGDDLYNMDTKESSTDVNRKILHTHGFKFLDRAAIISSRDHDNQKG 291
+ A + K+++PSN +L+ + R++ HT +FLD+AAI +
Sbjct: 258 FIVAFKSRKLQLPSNPSELHEFQMEHCVIRGKRQVYHTPTLRFLDKAAI-------KEDP 416
Query: 292 TCSNPWRL-CPVSQVEEVKCILRLLPIWLCTIIYSVVFTQMASLFVEQGDAMKTTVY-NF 349
T + R+ V+QVE K IL +L IWL T+I S ++ Q+ +LFV+QG + + +F
Sbjct: 417 TTGSSRRVPMTVNQVEGAKLILGMLLIWLVTLIPSTIWAQINTLFVKQGTTLDRNLGPDF 596
Query: 350 RVPPASMSCFDIISVAVFIFFYRRILDPFVGKLKKSDSKGLTELQRMGVGLVIAVIAMVS 409
++P AS+ F +S+ + + Y R+ PF+ + K +G+T LQR+G+G I +IA+
Sbjct: 597 KIPAASLGSFVTLSMLLSVPMYDRLFVPFM-RQKTGHPRGITLLQRLGIGFSIQIIAIAI 773
Query: 410 AGIVESYRLKHADQDCRHCKGSSTL---SIFWQIPQYALIGASEVFMYVGQLEFFNAQTP 466
A VE R+ ++ H G + SIFW +PQY LIG ++VF +G LEFF Q+P
Sbjct: 774 AYAVEVRRMHVIKEN--HIFGPKDIVPMSIFWLLPQYVLIGIADVFNAIGLLEFFYDQSP 947
Query: 467 DGLKSFGSALCMTSISLGNYVSSLLVSVVMKISTEDHMPGWIPVNLNKGHLDRFY 521
+ ++S G+ + I +GN+++S LV++ KI+ WI NLN HL +Y
Sbjct: 948 EDMQSLGTTFFTSGIGVGNFLNSFLVTMTDKITGRGDRKSWIADNLNDSHLXYYY 1112
>TC93408 similar to GP|11933407|dbj|BAB19758. putative nitrate transporter
NRT1-3 {Glycine max}, partial (44%)
Length = 909
Score = 200 bits (509), Expect = 1e-51
Identities = 112/303 (36%), Positives = 181/303 (58%), Gaps = 3/303 (0%)
Frame = +1
Query: 177 SNTILVYFEDEGMWALGFWLSAGCAFIALILFLVGTPKYRHFKPSGNPLARFCQVLVAAS 236
+NT+LVY +D W LG+ L I++++FL GTP YRH P+G+ R +V+VA+
Sbjct: 1 ANTVLVYIQDNVGWTLGYALPTLGLAISIMIFLGGTPFYRHKLPAGSTFTRMARVIVASL 180
Query: 237 RKVKVEMPSNGDDLYNMDTKESSTDVNR--KILHTHGFKFLDRAAIISSRDHDNQKGTCS 294
RK KV +P + LY +D +E + + +I T +FLD+A++ K +
Sbjct: 181 RKWKVPVPDDTKKLYELDMEEYAKKGSNTYRIDSTPTLRFLDKASV---------KTGST 333
Query: 295 NPWRLCPVSQVEEVKCILRLLPIWLCTIIYSVVFTQMASLFVEQGDAMKTTVYNFRVPPA 354
+PW LC V+ VEE K +LR++PI + T + S + Q+ +LFV+QG + + +F++PPA
Sbjct: 334 SPWMLCTVTHVEETKQMLRMIPILVATFVPSTMMAQVNTLFVKQGTTLDRHIGSFKIPPA 513
Query: 355 SMSCFDIISVAVFIFFYRRILDPFVGKLKKSDSKGLTELQRMGVGLVIAVIAMVSAGIVE 414
S++ F +S+ V + Y R + +L K + +G+T LQRMG+GLV+ I MV A + E
Sbjct: 514 SLAAFVTLSLLVCVVLYDRFFVRIMQRLTK-NPRGITLLQRMGIGLVLHTIIMVVASVTE 690
Query: 415 SYRLKHA-DQDCRHCKGSSTLSIFWQIPQYALIGASEVFMYVGQLEFFNAQTPDGLKSFG 473
+YRL+ A + G LSIF +PQ+ L+G ++ F+ V ++EFF Q P +KS G
Sbjct: 691 NYRLRVAKEHGLVESGGQVPLSIFILLPQFILMGTADAFLEVAKIEFFYDQAPXSMKSIG 870
Query: 474 SAL 476
+++
Sbjct: 871 TSI 879
>TC83116 similar to GP|11933407|dbj|BAB19758. putative nitrate transporter
NRT1-3 {Glycine max}, partial (50%)
Length = 1192
Score = 192 bits (489), Expect = 2e-49
Identities = 92/246 (37%), Positives = 157/246 (63%), Gaps = 1/246 (0%)
Frame = +3
Query: 13 VNQCLATLAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGR 72
V + +A++GI NL++ T+ + Q A NNV+ W GT+++ ++GA+++D++ GR
Sbjct: 165 VYEVFERMAYYGISSNLILLFTKKLHQGTXTAXNNVTNWVGTIWMTPILGAYVADAHLGR 344
Query: 73 YKTCAVFQVIFVIGLMSLSLSSYLFLIKPKGCGNESI-QCGKHSSWEMGMFYLSIYLIAL 131
+ T + I++ G+ L+L+ L +KP C + +C K S+ ++ +FY ++Y + +
Sbjct: 345 FWTFLIASFIYLSGMSLLTLAVSLPTLKPPECHELXVTKCKKLSTLQLAVFYGALYTLXI 524
Query: 132 GNGGYQPNIATFGADQFDEEHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGMWA 191
G GG +PNI+T GADQFD+ H KE +K++FF+++ ++ FG LF+NT+LVY +D W
Sbjct: 525 GTGGTKPNISTIGADQFDDFHPKEKSHKLSFFNWWMFSIFFGTLFANTVLVYIQDNVGWT 704
Query: 192 LGFWLSAGCAFIALILFLVGTPKYRHFKPSGNPLARFCQVLVAASRKVKVEMPSNGDDLY 251
LG+ L I++++FL GTP YRH P+G+ R +V+VA+ RK KV +P + LY
Sbjct: 705 LGYALPTLGLAISIMIFLGGTPFYRHKLPAGSTFTRMARVIVASLRKWKVPVPDDTKKLY 884
Query: 252 NMDTKE 257
+D +E
Sbjct: 885 ELDMEE 902
>BM780227 similar to PIR|E84798|E84 probable peptide/amino acid transporter
[imported] - Arabidopsis thaliana, partial (36%)
Length = 750
Score = 177 bits (448), Expect = 1e-44
Identities = 94/254 (37%), Positives = 154/254 (60%)
Frame = +3
Query: 28 NLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGRYKTCAVFQVIFVIGL 87
+LV++L V+ Q+ AA NV+ W G + L G F++D+Y GRY +++++GL
Sbjct: 3 SLVLYLILVIHQDLKTAARNVNYWAGVTTLMPLFGGFIADAYLGRYSAVVASSIVYLMGL 182
Query: 88 MSLSLSSYLFLIKPKGCGNESIQCGKHSSWEMGMFYLSIYLIALGNGGYQPNIATFGADQ 147
+ L+LS +L +KP C + + C + +F+L+IYLI++ GG++P++ +FGADQ
Sbjct: 183 ILLTLSWFLPSLKP--C-DHTTTCNEPRKIHEVVFFLAIYLISIATGGHKPSLESFGADQ 353
Query: 148 FDEEHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGMWALGFWLSAGCAFIALIL 207
FDE+H +E K++FF+++ AL G + T++VY +D W + G ++L++
Sbjct: 354 FDEDHVEERKQKMSFFNWWNCALCSGLILGVTLIVYIQDNINWGAADIIFTGVMALSLLI 533
Query: 208 FLVGTPKYRHFKPSGNPLARFCQVLVAASRKVKVEMPSNGDDLYNMDTKESSTDVNRKIL 267
F++G P YR+ PSG+PL QVLVAA K K+ PSN D LY + +S + + +
Sbjct: 534 FIIGRPFYRYRVPSGSPLTPMLQVLVAAFSKRKLPYPSNPDQLY--EVSKSHGNKRKFLC 707
Query: 268 HTHGFKFLDRAAII 281
HT +FL +AAII
Sbjct: 708 HTKKLRFL*QAAII 749
>BE997589 weakly similar to PIR|G86449|G86 hypothetical protein AAF81343.1
[imported] - Arabidopsis thaliana, partial (17%)
Length = 517
Score = 156 bits (395), Expect = 2e-38
Identities = 78/144 (54%), Positives = 99/144 (68%)
Frame = +2
Query: 130 ALGNGGYQPNIATFGADQFDEEHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGM 189
A G GG+QP +ATFGADQ+DE + KE KVAFF YFY +LN G LFSNT+LVY+ED G
Sbjct: 2 AFGYGGHQPTLATFGADQYDERNPKERSLKVAFFCYFYFSLNVGSLFSNTVLVYYEDTGK 181
Query: 190 WALGFWLSAGCAFIALILFLVGTPKYRHFKPSGNPLARFCQVLVAASRKVKVEMPSNGDD 249
W +GF++S A IAL+ FL G+PKYR+ KPSGNP+ R QV AA+RK V P+ D
Sbjct: 182 WTMGFFISLISAIIALLTFLSGSPKYRYLKPSGNPVVRVAQVFTAAARKWDV-APAKADK 358
Query: 250 LYNMDTKESSTDVNRKILHTHGFK 273
L+ + S+ RKILH+ F+
Sbjct: 359 LFEVLGSRSAIKGCRKILHSDDFR 430
>BI271961 similar to PIR|T47573|T475 peptide transport-like protein -
Arabidopsis thaliana, partial (12%)
Length = 680
Score = 147 bits (370), Expect = 1e-35
Identities = 80/230 (34%), Positives = 130/230 (55%), Gaps = 1/230 (0%)
Frame = +1
Query: 85 IGLMSLSLSSYLFLIKPKGCGNESIQCGKHSSWEMGMFYLSIYLIALGNGGYQPNIATFG 144
+G++ L++S+ ++P+ C + C + S ++ Y ++ LIALG GG +P +++FG
Sbjct: 1 VGMVLLTMSASFDTLRPQKCLAKP--CQEASQAQISFLYGALGLIALGTGGIKPCVSSFG 174
Query: 145 ADQFDEEHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGMWALGFWLSAGCAFIA 204
ADQFDE KE K AFF++F+ A+N G L TILVY +D+ W GF + ++
Sbjct: 175 ADQFDEGDEKEVQKKYAFFNWFFFAINMGALLGITILVYAQDKLGWGWGFGIPTITTVLS 354
Query: 205 LILFLVGTPKYRHFKPSGNPLARFCQVLVAASRKVKVEMP-SNGDDLYNMDTKESSTDVN 263
+++ G YR KP G+P RF QV+VA+ +K + + N LY ++T S
Sbjct: 355 IVVLAAGVRYYRFQKPMGSPFTRFLQVIVASVKKHQRGVSVXNEPTLYXVETTHSDIIGA 534
Query: 264 RKILHTHGFKFLDRAAIISSRDHDNQKGTCSNPWRLCPVSQVEEVKCILR 313
RK+ HT ++F D+A +I+ +D T N W +C +QVE K ++
Sbjct: 535 RKLPHTRQYRFFDKAXVITEKD------TLXNRWSVCTXTQVEXFKSFIK 666
>TC86048 weakly similar to PIR|H96561|H96561 probable peptide transporter
[imported] - Arabidopsis thaliana, partial (47%)
Length = 1364
Score = 144 bits (363), Expect = 9e-35
Identities = 91/315 (28%), Positives = 155/315 (48%), Gaps = 2/315 (0%)
Frame = +3
Query: 14 NQCLATLAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGRY 73
N+ LA +A G+G N++++L + A A + + LV AF++DSY GR+
Sbjct: 180 NEALARMASTGLGPNMILYLMGSYNLHLATATQILLISSAAGNFTPLVAAFIADSYIGRF 359
Query: 74 KTCAVFQVIFVIGLMSLSLSSYLFLIKPKGCGNESIQCGKHSSWEMGMFYLSIYLIALGN 133
+ +I +G+ L L++ + +P C N C +M M ++ LI +GN
Sbjct: 360 LGVGIGSIISFLGMAMLWLTAMVPAAQPPACSNPPEGCISAKPGQMAMLLSALILIGIGN 539
Query: 134 GGYQPNIATFGADQFD-EEHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGMWAL 192
GG ++A FGADQ +E+S FF+++Y + + + +VY +D W +
Sbjct: 540 GGISCSLA-FGADQVKRKENSNNNRALEIFFTWYYASTTIAVIAALFGIVYIQDHLGWRI 716
Query: 193 GFWLSAGCAFIALILFLVGTPKYRHFKPSGNPLARFCQVLVAASRKVKVEMPSNGDDLYN 252
GF + A I+ +LF + +P Y + L F QV VAA + K+ +P +
Sbjct: 717 GFGVPAALMLISTVLFFLASPLYVKITQRTSLLTGFAQVSVAAFKNRKLSLPPKTSPEFY 896
Query: 253 MDTKESSTDVNRKILHTHGFKFLDRAAIISSRDHD-NQKGTCSNPWRLCPVSQVEEVKCI 311
K+S ++ T +FL++A +I + D G+ N W LC V QVEE+K I
Sbjct: 897 HHKKDSVL-----VVPTDKLRFLNKACVIKDHEQDIASDGSIINRWSLCTVDQVEELKAI 1061
Query: 312 LRLLPIWLCTIIYSV 326
++++P+W I S+
Sbjct: 1062IKVIPLWSTGITMSI 1106
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.327 0.141 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,571,595
Number of Sequences: 36976
Number of extensions: 307075
Number of successful extensions: 2157
Number of sequences better than 10.0: 116
Number of HSP's better than 10.0 without gapping: 2052
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2073
length of query: 561
length of database: 9,014,727
effective HSP length: 101
effective length of query: 460
effective length of database: 5,280,151
effective search space: 2428869460
effective search space used: 2428869460
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144516.1


