

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144515.12 - phase: 0
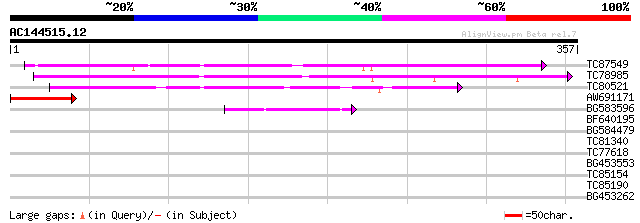
(357 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87549 similar to PIR|T01734|T01734 hypothetical protein A_IG00... 125 2e-29
TC78985 similar to PIR|T06001|T06001 hypothetical protein F17M5.... 115 2e-26
TC80521 weakly similar to GP|18377712|gb|AAL67006.1 unknown prot... 89 3e-18
AW691171 73 1e-13
BG583596 similar to GP|4325376|gb| T3H13.12 gene product {Arabid... 42 4e-04
BF640195 similar to GP|13374856|em putative protein {Arabidopsis... 36 0.020
BG584479 similar to GP|9758299|dbj| gene_id:MJH22.4~pir||T01734~... 32 0.49
TC81340 similar to PIR|A85296|A85296 hypothetical protein AT4g25... 28 5.4
TC77618 homologue to PIR|S57478|S57478 GTP-binding protein GTP13... 28 5.4
BG453553 weakly similar to GP|15341853|gb Unknown (protein for M... 27 9.3
TC85154 similar to GP|8894548|emb|CAB95829.1 hypothetical protei... 27 9.3
TC85190 similar to GP|8894548|emb|CAB95829.1 hypothetical protei... 27 9.3
BG453262 homologue to GP|22655172|gb| unknown protein {Arabidops... 27 9.3
>TC87549 similar to PIR|T01734|T01734 hypothetical protein A_IG002N01.30 -
Arabidopsis thaliana (fragment), partial (38%)
Length = 1766
Score = 125 bits (315), Expect = 2e-29
Identities = 97/337 (28%), Positives = 164/337 (47%), Gaps = 8/337 (2%)
Frame = +2
Query: 10 RFDHIRTFVNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKAS 69
RF+ I ++A VK ++ D + K+K LK V+ ++ +VS P++ +SL +
Sbjct: 422 RFNKIMP-ISAAVKRRKELPFDNVIQKDKKLKFVLKVRKILVSKPTRVMSLQELGKYRRE 598
Query: 70 LNLPMIT--IKLVEKYHHVFMQFQPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDAA 127
L L I ++ ++ VF + + K+T++A L+ EE+ V N ++
Sbjct: 599 LGLDKKRKLIVILRRFPGVF-EIVEDGCYSLKFKMTSEAEKLYLEELRVRNEM--EDVVV 769
Query: 128 QRLARLLMLAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVL 187
+L +LLM+ R+ + I L D+GLP ++ T+ YP++F V E
Sbjct: 770 TKLRKLLMMPLEKRILLEKIGHLANDLGLPREFRDTICHRYPEFFKVVQTE------RGP 931
Query: 188 ALELVSWRKELSVSELEKRVMGFNYDADKRRHDI--QFPMF----LPKSFALEKRVKTWV 241
ALEL W L+VS E + ++ I + P F LPK +L K +
Sbjct: 932 ALELTHWDPHLAVSAAELSAEENRIREVEEQNLIIDRAPKFNRVKLPKGLSLSKGEMRKI 1111
Query: 242 EGWQTLPYISPYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLG 301
++ +PYISPY + + N+ + EK ++HE+LSL + K+T +N + E
Sbjct: 1112MQFRDIPYISPYSDFSMIGLNTPEKEKHACGVIHEILSLTLEKRTLVDNFTHFREEFRFS 1291
Query: 302 SRFKKALVHHPGIFYLSNKIRTQTAVLREAYRNEFLV 338
+ + L+ HP +FY+S K + LREAYR+ LV
Sbjct: 1292QQLRGMLIRHPDMFYISLKGDRDSVFLREAYRDSQLV 1402
>TC78985 similar to PIR|T06001|T06001 hypothetical protein F17M5.260 -
Arabidopsis thaliana, partial (33%)
Length = 2496
Score = 115 bits (289), Expect = 2e-26
Identities = 95/350 (27%), Positives = 165/350 (47%), Gaps = 11/350 (3%)
Frame = +1
Query: 16 TFVNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNLPMI 75
T + + + +RD D + EK + V+ +N I+S P++S+ + +
Sbjct: 220 TSIPKKQQRIRDHGYDNYMEIEKKTRKVLKFQNLILSEPNQSIPIPRLETQAHRIGFTRH 399
Query: 76 -TIKLVEKYHHVFMQFQPSPGLPPRIKLTAQA-LALHKEEMEVHNSRTNQEDAAQRLARL 133
I + K+ HVF F+ +LT +A L + +E + + + E A RL ++
Sbjct: 400 EAIAFILKFPHVFEIFEHPVQRILFCRLTRKAILQIEQERLALADQ---VERAVTRLRKM 570
Query: 134 LMLAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALELVS 193
+M++ RL + + + +GLP D+ +++ YP++F + ++ + +ELV
Sbjct: 571 IMMSNGLRLRLEHVRIARSALGLPDDFEYSVVLRYPEFFRLVDAKETRNKY----IELVE 738
Query: 194 WRKELSVSELEKRVMGFNYDADKRRHDIQFPMFL--PKSFALEKRVKTWVEGWQTLPYIS 251
+ +L+ +E + DI+F + P F + K + + WQ LPY S
Sbjct: 739 FDPKLAKCAIEDARERVYRERGSEAEDIRFSFLIDFPPGFKISKYFRIAMWKWQRLPYWS 918
Query: 252 PYENAFHLDSNSDQA----EKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKA 307
PYE+ D S +A EK VA +HELLSL V KK E + + + L + K+
Sbjct: 919 PYEDVLGYDLRSIEAQKRMEKRAVATIHELLSLTVEKKITLERIAHFRMAMNLPKKLKEI 1098
Query: 308 LVHHPGIFYLS---NKIRTQTAVLREAYRNEFLVKNHPVMGTRYWYIHLM 354
L+ H GIFY+S N + T LREAYR LV+ + + R + L+
Sbjct: 1099LLQHQGIFYVSTRGNHGKLHTVFLREAYRKGELVEPNDLYLARRKLVELV 1248
>TC80521 weakly similar to GP|18377712|gb|AAL67006.1 unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 1025
Score = 89.0 bits (219), Expect = 3e-18
Identities = 72/262 (27%), Positives = 123/262 (46%), Gaps = 2/262 (0%)
Frame = +2
Query: 26 RDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNLPMITIKLVEKYHH 85
+D L++A+ + K +KN I+ P+ + + +L+L + + KY
Sbjct: 296 KDPDLESALSRNKRWIINNQIKNIILRYPNNQIPIQTLQKKFKTLDLQGKALNWISKYPS 475
Query: 86 VFMQFQPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLARLLMLAGMARLPIY 145
F Q + LT + + L EE + +S + RLA+LLML+ L +
Sbjct: 476 CFQFHQD------HVLLTKRMMELVHEEQSLKDSL--ESVFVPRLAKLLMLSLNNCLNVM 631
Query: 146 VIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALELVSWRKELSVSELEK 205
I ++K +G P DY+ ++A YPD F + + + + +EL+ W + +VSE+E
Sbjct: 632 KINEIKNSLGFPDDYLIGIVAKYPDLFR---IRNESGRRSSMVVELMKWNPDFAVSEVEA 802
Query: 206 RVMGFNYDADKRRHDIQFPMFLPKSF--ALEKRVKTWVEGWQTLPYISPYENAFHLDSNS 263
M K ++ F LP S+ +LEK ++ +PY+SPY + L S
Sbjct: 803 LAM-------KNGVEVNFSCCLPSSWVKSLEK-----FHEFELVPYVSPYSDPRGLVEGS 946
Query: 264 DQAEKWTVAILHELLSLLVSKK 285
+ EK V ++HELL L + KK
Sbjct: 947 KEMEKRNVGLVHELLXLTLWKK 1012
>AW691171
Length = 431
Score = 73.2 bits (178), Expect = 1e-13
Identities = 35/43 (81%), Positives = 40/43 (92%), Gaps = 1/43 (2%)
Frame = +1
Query: 1 MAQSTKLFHRFDHIRTFVNARVKWVR-DDYLDTAVLKEKNLKH 42
MA+STK+ +RFDHIRTFVNA+VKWVR DDYLDTAVLKEKNL +
Sbjct: 289 MARSTKIVYRFDHIRTFVNAKVKWVRDDDYLDTAVLKEKNLSY 417
>BG583596 similar to GP|4325376|gb| T3H13.12 gene product {Arabidopsis
thaliana}, partial (46%)
Length = 778
Score = 42.0 bits (97), Expect = 4e-04
Identities = 28/83 (33%), Positives = 45/83 (53%)
Frame = +3
Query: 136 LAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALELVSWR 195
L+ R+ + I ++ G+P D+ + YP+YF + ++ GK+V LELV+W
Sbjct: 456 LSRFHRISLAKIHHVRNLFGIPDDF-RDRVEKYPNYFRIVDDKNKDDGKKVRVLELVNWD 632
Query: 196 KELSVSELEKRVMGFNYDADKRR 218
L+VS +EK M N D KR+
Sbjct: 633 PLLAVSAIEKEFM-VNEDDAKRK 698
>BF640195 similar to GP|13374856|em putative protein {Arabidopsis thaliana},
partial (32%)
Length = 521
Score = 36.2 bits (82), Expect = 0.020
Identities = 41/164 (25%), Positives = 67/164 (40%), Gaps = 9/164 (5%)
Frame = +2
Query: 8 FHRFDHIRTFVNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLK 67
F R+ V R K R L+ A K K ++ L + P + + + S +
Sbjct: 50 FERWMTTSKRVQDRSKKKRVHELEAATEKWKITSKIVFLIELLEQEPEIVIPVRSLSHYR 229
Query: 68 ASLNLPMITIKLVEKYHHVFMQFQPSPGLPPRIK---------LTAQALALHKEEMEVHN 118
++LP K H + + +P L K LT +A L +E V
Sbjct: 230 KQISLP--------KPHRISDFLRKTPKLFELYKDRNGVLWCGLTQKAEVLMEEHKRVIE 385
Query: 119 SRTNQEDAAQRLARLLMLAGMARLPIYVIEKLKWDMGLPHDYVT 162
N++ AA+ + RLLM++ R+ + I + D GLP D+ T
Sbjct: 386 E--NEDKAAEYVTRLLMMSVDKRVQLDKIAHFRRDFGLPMDFTT 511
>BG584479 similar to GP|9758299|dbj| gene_id:MJH22.4~pir||T01734~similar to
unknown protein {Arabidopsis thaliana}, partial (16%)
Length = 786
Score = 31.6 bits (70), Expect = 0.49
Identities = 15/56 (26%), Positives = 31/56 (54%)
Frame = +1
Query: 277 LLSLLVSKKTESENLICYGECLGLGSRFKKALVHHPGIFYLSNKIRTQTAVLREAY 332
+L + + K+T ++L + + GL ++ + +V HP +FY S K + + L E +
Sbjct: 1 VLGMTIEKRTLIDHLTHFRKEFGLPNKLRGMIVRHPELFYTSLKGQRDSVFLVERF 168
>TC81340 similar to PIR|A85296|A85296 hypothetical protein AT4g25610
[imported] - Arabidopsis thaliana, partial (13%)
Length = 1419
Score = 28.1 bits (61), Expect = 5.4
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 2/64 (3%)
Frame = +2
Query: 9 HRFDHIRTFVNARVKWVRDDY--LDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASML 66
+R + VN+ W R +DT +LK K K +K H V S S++L N S
Sbjct: 608 NRDQRVAPIVNSGKVWSRKPKPEIDTVILKPKLHKEPDEVKTHEVLIGSVSVTLVNCSQS 787
Query: 67 KASL 70
+ +L
Sbjct: 788 ENNL 799
>TC77618 homologue to PIR|S57478|S57478 GTP-binding protein GTP13 - garden
pea, complete
Length = 1136
Score = 28.1 bits (61), Expect = 5.4
Identities = 27/85 (31%), Positives = 38/85 (43%), Gaps = 6/85 (7%)
Frame = -3
Query: 51 VSSPSKSLSLYNASMLKASLNLPMITIKLVEKYHHVFM---QFQPSPGLPPRIKLTA--- 104
VS + L ++SM+ ++PM+ IKLV K + Q P+P P R L
Sbjct: 429 VSQICNLMRLPSSSMVLILKSMPMVVIKLVVKEPSENLKSKQLFPTPLSPIRRSLMR*S* 250
Query: 105 QALALHKEEMEVHNSRTNQEDAAQR 129
A AL NS+ N + AQR
Sbjct: 249 SARALAGAAAIAQNSKFNTQSTAQR 175
>BG453553 weakly similar to GP|15341853|gb Unknown (protein for MGC:9496)
{Homo sapiens}, partial (81%)
Length = 653
Score = 27.3 bits (59), Expect = 9.3
Identities = 13/30 (43%), Positives = 20/30 (66%)
Frame = +3
Query: 51 VSSPSKSLSLYNASMLKASLNLPMITIKLV 80
+SSP K+ +L N+SML LN ++T L+
Sbjct: 246 LSSPLKAFTLVNSSMLAERLN*ALVTSSLL 335
>TC85154 similar to GP|8894548|emb|CAB95829.1 hypothetical protein {Cicer
arietinum}, partial (83%)
Length = 1811
Score = 27.3 bits (59), Expect = 9.3
Identities = 18/70 (25%), Positives = 31/70 (43%), Gaps = 15/70 (21%)
Frame = +3
Query: 190 ELVSWRKELSVSELEKRVMGFNYD-------ADKRRHDIQFPMF--------LPKSFALE 234
+ V WRKE V L + +G ++D DK H + + +F K+F+ E
Sbjct: 549 QTVLWRKEFGVEALLQEDLGTDWDKVVFTDGTDKEGHPVYYNVFGEFEDKDLYQKTFSDE 728
Query: 235 KRVKTWVEGW 244
++ +V W
Sbjct: 729 EKRTKFVRWW 758
>TC85190 similar to GP|8894548|emb|CAB95829.1 hypothetical protein {Cicer
arietinum}, partial (50%)
Length = 1496
Score = 27.3 bits (59), Expect = 9.3
Identities = 18/70 (25%), Positives = 31/70 (43%), Gaps = 15/70 (21%)
Frame = +3
Query: 190 ELVSWRKELSVSELEKRVMGFNYD-------ADKRRHDIQFPMF--------LPKSFALE 234
+ V WRKE V L + +G ++D DK H + + +F K+F+ E
Sbjct: 1005 QTVLWRKEFGVEALLQEDLGTDWDKVVFTDGTDKEGHPVYYNVFGEFEDKDLYQKTFSDE 1184
Query: 235 KRVKTWVEGW 244
++ +V W
Sbjct: 1185 EKRTKFVRWW 1214
>BG453262 homologue to GP|22655172|gb| unknown protein {Arabidopsis
thaliana}, partial (2%)
Length = 651
Score = 27.3 bits (59), Expect = 9.3
Identities = 19/73 (26%), Positives = 37/73 (50%), Gaps = 4/73 (5%)
Frame = +3
Query: 22 VKWVRDDYLDTAVLK----EKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNLPMITI 77
++++ D+L+ A + ++ K +I + H P+ SL Y + A + LP+I +
Sbjct: 30 IQYLSMDFLECANHRVHQASESRKSIIQQQQHGQPFPNLSLMAYLSQ*PDAEVKLPLINV 209
Query: 78 KLVEKYHHVFMQF 90
KLV + F+ F
Sbjct: 210 KLVLFGVNYFLTF 248
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,300,294
Number of Sequences: 36976
Number of extensions: 176998
Number of successful extensions: 916
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 909
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 910
length of query: 357
length of database: 9,014,727
effective HSP length: 97
effective length of query: 260
effective length of database: 5,428,055
effective search space: 1411294300
effective search space used: 1411294300
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144515.12


