

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144513.13 + phase: 0
(544 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
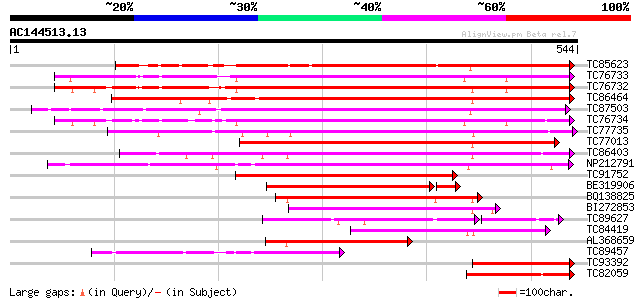
Score E
Sequences producing significant alignments: (bits) Value
TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 ... 409 e-114
TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 386 e-107
TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.... 384 e-107
TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Cit... 379 e-105
TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precurs... 375 e-104
TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 374 e-104
TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectineste... 373 e-104
TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylestera... 365 e-101
TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF028... 365 e-101
NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1 302 2e-82
TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectineste... 246 2e-65
BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precurso... 212 2e-59
BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like pr... 205 4e-53
BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-... 179 2e-45
TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pec... 137 5e-45
TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor ... 173 2e-43
AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886... 169 3e-42
TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE... 159 3e-39
TC93392 similar to PIR|T49241|T49241 pectinesterase-like protein... 138 6e-33
TC82059 similar to GP|19424045|gb|AAL87311.1 unknown protein {Ar... 135 5e-32
>TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 {Arabidopsis
thaliana}, partial (79%)
Length = 2442
Score = 409 bits (1052), Expect = e-114
Identities = 222/449 (49%), Positives = 291/449 (64%), Gaps = 8/449 (1%)
Frame = +3
Query: 102 REQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCIEG 161
+E+ A DC EL DF+V +L+ + + QYE + WLS AL+N +TC G
Sbjct: 837 QEKAAWADCLELYDFTVQKLSQT--------KYTKCTQYES--QTWLSTALTNLETCKNG 986
Query: 162 FEGTDRRLESYISGSVTQ-VTQLISNVLSLYTQLNRLPFRPPRNTTLHETSTDESLEFPE 220
F D + +Y+ ++ VT+L+SN LSL N++P++ P FP
Sbjct: 987 FY--DLGVTNYVLPLLSNNVTKLLSNTLSL----NKVPYQQPSYKD----------GFPT 1118
Query: 221 WMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKE 280
W+ D++LL++ A+ VVA DGSG+Y T+ A +AAPS S R +VIYVK G+Y E
Sbjct: 1119 WVKPGDRKLLQTSSAASKANVVVAKDGSGKYTTVKAATDAAPSGSGR-YVIYVKAGVYNE 1295
Query: 281 NIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAG 340
+++K K N+M+VGDGIG+TI+T +++ G TTFR+AT A +G GFIA+D+TFRNTAG
Sbjct: 1296 QVEIKAK--NVMLVGDGIGKTIITGSKSVGGGTTTFRSATVAATGDGFIAQDITFRNTAG 1469
Query: 341 PVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQ 400
NHQAVA R SD S F++CS EG QDTLY HS RQFYREC IYGT+DFIFGN A VLQ
Sbjct: 1470 AANHQAVAFRSGSDLSVFYKCSFEGYQDTLYVHSERQFYRECNIYGTVDFIFGNAAVVLQ 1649
Query: 401 NCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLA-------SQPTYLGRPWKEY 453
NC I+ R P P + +T+TAQGR P+Q+TG I +S V A S +YLGRPW++Y
Sbjct: 1650 NCNIFARNP-PAKTITVTAQGRTDPNQNTGIIIHNSRVSAQSDLNPSSVKSYLGRPWQKY 1826
Query: 454 SRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDA 513
SRTV++ T + + P GWL W GNFALDTL+Y EY N G GSS + RV W GYHV+ A
Sbjct: 1827 SRTVFMKTVLDGFINPAGWLPWDGNFALDTLYYAEYANTGSGSSTSNRVTWKGYHVLTSA 2006
Query: 514 SAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
S A FTV F+ G SW+ TGV FT+GL
Sbjct: 2007 SQASPFTVGNFIAGNSWIGNTGVPFTSGL 2093
>TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, partial (98%)
Length = 2029
Score = 386 bits (992), Expect = e-107
Identities = 216/517 (41%), Positives = 305/517 (58%), Gaps = 18/517 (3%)
Frame = +2
Query: 44 CMDIENQNSCLTNI----HNELTRTGPPSPTSVINAALRTTINEAIGAINNMTKISTFSV 99
C + SCL ++ H T V+ + L + A++ + I
Sbjct: 212 CEHAVDTKSCLAHVSEVSHVPTLVTTKDQNLHVLLSLLTKSTTHIQNAMDTASVIKRRIN 391
Query: 100 NNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCI 159
+ RE++A+ DC++L+D S++ + W M ++ + +Q + + WLS+ L+N TC+
Sbjct: 392 SPREEIALSDCEQLMDLSMNRI-WDT--MLKLTKNNIDSQQDAH--TWLSSVLTNHATCL 556
Query: 160 EGFEGTDRR-LESYISGSVTQVTQLISNVLSLYTQLNRLPFRPPRNTTLHETSTDESL-- 216
+G EG+ R +E+ + +++ ++ L ++ Q +R F DE+L
Sbjct: 557 DGLEGSSRVVMENDLQDLISRARSSLAVFLVVFPQKDRDQF------------IDETLIG 700
Query: 217 EFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKG 276
EFP W+T D+ LL++ A+ VVA DGSG+++T+ EAV +AP + ++VIYVKKG
Sbjct: 701 EFPSWVTSKDRRLLETAVGDIKANVVVAQDGSGKFKTVAEAVASAPDNGKTKYVIYVKKG 880
Query: 277 LYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFR 336
YKEN+++ K TN+M+VGDG+ TI+T N NF+ G TTF+++T A G GFIA+D+ F+
Sbjct: 881 TYKENVEIGSKKTNVMLVGDGMDATIITGNLNFIDGTTTFKSSTVAAVGDGFIAQDIWFQ 1060
Query: 337 NTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGA 396
N AG HQAVALRV SDQS RC I+ QDTLYAHS RQFYR+ I GTIDFIFGN A
Sbjct: 1061NMAGAAKHQAVALRVGSDQSVINRCRIDAFQDTLYAHSNRQFYRDSVITGTIDFIFGNAA 1240
Query: 397 AVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQD---------SYVLASQPTYLG 447
V Q CK+ R P+ Q TAQGR+ P Q+TG +IQ V+ S T+LG
Sbjct: 1241VVFQKCKLVARKPMANQNNMFTAQGREDPGQNTGTSIQQCDLTPSSDLKPVVGSIKTFLG 1420
Query: 448 RPWKEYSRTVYINTYMSSMVQPRGWLEW--LGNFALDTLWYGEYRNYGPGSSLAGRVKWP 505
RPWK+YSRTV + +++ S + P GW EW L TL+YGEY N GPG+ A RV WP
Sbjct: 1421RPWKKYSRTVVMQSFLDSHIDPTGWAEWDAASKDFLQTLYYGEYLNNGPGAGTAKRVTWP 1600
Query: 506 GYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
GYHVI A+ A FTV + + G WL TGV FT GL
Sbjct: 1601GYHVINTAAEASKFTVAQLIQGNVWLKNTGVAFTEGL 1711
>TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11)
precursor - garden pea, complete
Length = 1860
Score = 384 bits (986), Expect = e-107
Identities = 217/521 (41%), Positives = 316/521 (60%), Gaps = 22/521 (4%)
Frame = +2
Query: 44 CMDIENQNSCLTNIH-----NELTRTGPPSPTSVINAALRTT--INEAIGAINNMTKIST 96
C + SCLT++ + L+ T +++++ ++T I +A+ N + +
Sbjct: 197 CEHALDTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKR--- 367
Query: 97 FSVNN-REQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQ 155
VN+ RE++A+ DC+EL+D S+ + W + + + +Q++ + WLS+ L+N
Sbjct: 368 -RVNSPREEIALNDCEELMDLSMDRV-WD--SVLTLTKNNIDSQHDAH--TWLSSVLTNH 529
Query: 156 DTCIEGFEGTDRR-LESYISGSVTQVTQLISNVLSLYTQLNRLPFRPPRNTTLHETSTDE 214
TC++G EG+ R +ES + +++ ++ ++S+ PP+ ++ DE
Sbjct: 530 ATCLDGLEGSSRVVMESDLHDLISRARSSLAVLVSVL---------PPK---ANDGFIDE 673
Query: 215 SL--EFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIY 272
L +FP W+T D+ LL+S A+ VVA DGSG+++T+ +AV +AP + R+VIY
Sbjct: 674 KLNGDFPSWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIY 853
Query: 273 VKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKD 332
VKKG YKENI++ KK TN+M+VGDG+ TI+T + NF+ G TTF++AT A G GFIA+D
Sbjct: 854 VKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQD 1033
Query: 333 MTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIF 392
+ F+NTAGP HQAVALRV +DQS RC I+ QDTLYAHS RQFYR+ I GT+DFIF
Sbjct: 1034IRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIF 1213
Query: 393 GNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLASQ---------P 443
GN A V Q K+ R P+ QK +TAQGR+ P+Q+T +IQ V+ S
Sbjct: 1214GNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIK 1393
Query: 444 TYLGRPWKEYSRTVYINTYMSSMVQPRGWLEW--LGNFALDTLWYGEYRNYGPGSSLAGR 501
TYLGRPWK+YSRTV + + + + P GW EW L TL+YGEY N G G+ R
Sbjct: 1394TYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKR 1573
Query: 502 VKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
V WPGYH+IK+A+ A FTV + + G WL TGV F GL
Sbjct: 1574VTWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 1696
>TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Citrus
sinensis}, partial (85%)
Length = 1993
Score = 379 bits (972), Expect = e-105
Identities = 202/467 (43%), Positives = 286/467 (60%), Gaps = 22/467 (4%)
Frame = +1
Query: 98 SVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDT 157
S+ RE++A+ DC E +D ++ EL + ++ + Q+ +L+ +S+A++NQ T
Sbjct: 430 SLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVT 609
Query: 158 CIEGF--EGTDRRLESYISGSVTQVTQLISNVLSL-----------YTQLNRLPFRPPRN 204
C++GF + D+ + + V + SN L++ + Q N +
Sbjct: 610 CLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNR 789
Query: 205 TTLHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSH 264
L E + + +PEW++ D+ LL+ AD VVA DGSG ++T++EAV AAP
Sbjct: 790 KLLEE---ENGVGWPEWISAGDRRLLQGSTVK--ADVVVAADGSGNFKTVSEAVAAAPLK 954
Query: 265 SNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVS 324
S++R+VI +K G+YKEN+++ KK TNIM +GDG TI+T +RN + G TTF +AT A+
Sbjct: 955 SSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIV 1134
Query: 325 GKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEI 384
G F+A+D+TF+NTAGP HQAVALRV +D SAF+ C I QDTLY H+ RQF+ C I
Sbjct: 1135GGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFI 1314
Query: 385 YGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLASQ-- 442
GT+DFIFGN A V QNC I+ R P QK +TAQGR P+Q+TG IQ + A++
Sbjct: 1315SGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDL 1494
Query: 443 -------PTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPG 495
PTYLGRPWKEYSRTV++ + +S ++ P GW EW GNFAL+TL Y EY+N GPG
Sbjct: 1495EGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPG 1674
Query: 496 SSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
+ + RV W G+ VI A+ A T F+ G SWL TG F+ GL
Sbjct: 1675AGTSKRVTWKGFKVITSAAEAQSSTPGNFIGGSSWLGSTGFPFSLGL 1815
>TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precursor (EC
3.1.1.11) (Pectin methylesterase 3) (PE 3)., partial
(93%)
Length = 2088
Score = 375 bits (962), Expect = e-104
Identities = 215/529 (40%), Positives = 306/529 (57%), Gaps = 12/529 (2%)
Frame = +2
Query: 22 SFPDEIPSDIQTQDMQALIAQACMDIENQNSCLTNIHNELTRTGPPSPTSVINAALRTTI 81
S P+ +P+ T + C + NSC ++I + L + P + +L+ I
Sbjct: 239 SSPNSLPNTELTPATS--LKAVCESTQYPNSCFSSI-SSLPDSNTTDPEQLFKLSLKVAI 409
Query: 82 NEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYE 141
+E ++T+ S + R + AI C +L S+ L S+ + + G + +
Sbjct: 410 DELSKL--SLTRFSEKATEPRVKKAIGVCDNVLADSLDRLNDSMSTI--VDGGKMLSPAK 577
Query: 142 -GNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQV----TQLISNVLSLYTQLNR 196
++E WLSAAL++ DTC++ + + + ++ T+ SN L++ +++ R
Sbjct: 578 IRDVETWLSAALTDHDTCLDAVGEVNSTAARGVIPEIERIMRNSTEFASNSLAIVSKVIR 757
Query: 197 LPFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINE 256
L N + EFPEW+ A++ LL + + + DAVVA DGSGQY+TI E
Sbjct: 758 LL----SNFEVSNHHRRLLGEFPEWLGTAERRLLATVVNETVPDAVVAKDGSGQYKTIGE 925
Query: 257 AVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTF 316
A+ S +R V+YVKKG+Y ENID+ K N+M+ GDG+ +T+V+ +RN++ G TF
Sbjct: 926 ALKLVKKKSLQRFVVYVKKGVYVENIDLDKNTWNVMIYGDGMTETVVSGSRNYIDGTPTF 1105
Query: 317 RTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLR 376
TATFAV GKGFIAKD+ F NTAG HQAVA+R SDQS F+RCS G QDTLYAHS R
Sbjct: 1106ETATFAVKGKGFIAKDIQFLNTAGASKHQAVAMRSGSDQSVFYRCSFVGYQDTLYAHSNR 1285
Query: 377 QFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDS 436
QFYR+C+I GTIDFIFGN AAV QNCKI R P+ Q TITAQG+K P+Q++G IQ S
Sbjct: 1286QFYRDCDITGTIDFIFGNAAAVFQNCKIMPRQPMSNQFNTITAQGKKDPNQNSGIVIQKS 1465
Query: 437 YVLA------SQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNF-ALDTLWYGEY 489
PTYLGRPWK++S T+ + + + S ++P GW+ W+ N ++ Y EY
Sbjct: 1466TFTTLPGDNLIAPTYLGRPWKDFSTTIIMKSEIGSFLKPVGWISWVANVEPPSSILYAEY 1645
Query: 490 RNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKF 538
+N GPG+ + GRVKW GY A FTV F+ G WLP V+F
Sbjct: 1646QNTGPGADVPGRVKWAGYKPALGDEDAIKFTVDSFIQGPEWLPSASVQF 1792
>TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, complete
Length = 1981
Score = 374 bits (959), Expect = e-104
Identities = 217/520 (41%), Positives = 309/520 (58%), Gaps = 21/520 (4%)
Frame = +3
Query: 44 CMDIENQNSCLTNIH-----NELTRTGPPSPTSVINAALRTT--INEAIGAINNMTKIST 96
C + NSCLT++ + L T +++I+ ++T I +A+ N + +
Sbjct: 204 CEHAVDTNSCLTHVAEVVQGSTLDNTKDHKLSTLISLLTKSTTHIRKAMDTANVIKR--- 374
Query: 97 FSVNN-REQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQ 155
+N+ RE+ A+ C++L++ S+ E W + + + +Q + + WLS+ L+N
Sbjct: 375 -RINSPREENALNVCEKLMNLSM-ERVWD--SVLTLTKDNMDSQQDAH--TWLSSVLTNH 536
Query: 156 DTCIEGFEGTDRRLESYISGSVTQVTQLISNVLSLYTQLNRLPFRPPRNTTLHETSTDES 215
TC++G EGT R + + LI+ S L + PP++ H+ DES
Sbjct: 537 ATCLDGLEGTSRAVME------NDIQDLIARARSSLAVL--VAVLPPKD---HDEFIDES 683
Query: 216 L--EFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYV 273
L +FP W+T D+ LL+S A+ VVA DGSG+++T+ EAV +AP+ R+VIYV
Sbjct: 684 LNGDFPSWVTSKDRRLLESSVGDVKANVVVAKDGSGKFKTVAEAVASAPNKGTARYVIYV 863
Query: 274 KKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDM 333
KKG+YKEN+++ TN+M++GDG+ TI+T + N++ G TF+TAT A G FIA+D+
Sbjct: 864 KKGIYKENVEIASSKTNVMLLGDGMDATIITGSLNYVDGTGTFQTATVAAVGDWFIAQDI 1043
Query: 334 TFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFG 393
F+NTAGP HQAVALRV SD+S RC I+ QDTLYAH+ RQFYR+ I GTIDFIFG
Sbjct: 1044GFQNTAGPQKHQAVALRVGSDRSVINRCKIDAFQDTLYAHTNRQFYRDSFITGTIDFIFG 1223
Query: 394 NGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQD---------SYVLASQPT 444
+ A VLQ CK+ R P+ Q +TAQGR P+Q+T +IQ V+ S T
Sbjct: 1224DAAVVLQKCKLVARKPMANQNNMVTAQGRIDPNQNTATSIQQCDVIPSTDLKPVIGSVKT 1403
Query: 445 YLGRPWKEYSRTVYINTYMSSMVQPRGWLEW--LGNFALDTLWYGEYRNYGPGSSLAGRV 502
YLGRPWK+YSRTV + + + + + P GW EW L TL+YGEY N GPG+ + RV
Sbjct: 1404YLGRPWKKYSRTVVMQSLLGAHIDPTGWAEWDAASKDFLQTLYYGEYMNSGPGAGTSKRV 1583
Query: 503 KWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
KWPGYH+I A A FTV + + G WL TGV F AGL
Sbjct: 1584KWPGYHIINTAE-ANKFTVAQLIQGNVWLKNTGVAFIAGL 1700
>TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectinesterase
{Arabidopsis thaliana}, partial (59%)
Length = 1963
Score = 373 bits (958), Expect = e-104
Identities = 204/488 (41%), Positives = 290/488 (58%), Gaps = 38/488 (7%)
Frame = +2
Query: 95 STFSVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYE--GNLEAWLSAAL 152
S+ S+N+ E A+ DCK+L +V L E++ + ++ E +E++LSA
Sbjct: 317 SSSSLNHEEVGALVDCKDLNSLNVDYLESISDELKSASSSSSSSDTELVDKIESYLSAVA 496
Query: 153 SNQDTCIEGFEGTDRRLESYISGSVTQVTQLISNVLSLYTQLNRLPFRPPRNTTLHETST 212
+N TC +G T + + ++ + TQ S L L T+ L RN T
Sbjct: 497 TNHYTCYDGLVVTKSNIANALAVPLKDATQFYSVSLGLVTEA--LSKNMKRNKTRKHGLP 670
Query: 213 DESLEFPEWM----------------------TEADQELLKSKPHGKIADAVVALD--GS 248
++S + + + T ++ L +S+ HG + + V + G
Sbjct: 671 NKSFKVRQPLEKLIKLLRTKYSCQKTSSNCTSTRTERILKESESHGILLNDFVLVSPYGI 850
Query: 249 GQYRTINEAVNAAPSHSNRR---HVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTS 305
+ +I +A+ AAP+++ ++IYV++G Y+E + + K NI++VGDGI TI+T
Sbjct: 851 ANHTSIGDAIAAAPNNTKPEDGYYLIYVREGYYEEYVIVPKHKNNILLVGDGINNTIITG 1030
Query: 306 NRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEG 365
N + + GWTTF ++TFAVSG+ FIA D+TFRNTAGP HQAVA+R ++D S F+RCS EG
Sbjct: 1031NHSVIDGWTTFNSSTFAVSGERFIAVDITFRNTAGPEKHQAVAVRNNADLSTFYRCSFEG 1210
Query: 366 NQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSP 425
QDTLY HS+RQFYR+C+IYGT+DFIFGN A V QNC IY R PLP QK +TAQGR P
Sbjct: 1211YQDTLYVHSVRQFYRDCKIYGTVDFIFGNAAVVFQNCNIYARKPLPNQKNAVTAQGRTDP 1390
Query: 426 HQSTGFTIQDSYVLASQP---------TYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWL 476
+Q+TG +IQ+ + A+Q +YLGRPWK YSRTVY+ +Y+ VQP GWLEW
Sbjct: 1391NQNTGISIQNCTIDAAQDLANDLNSTMSYLGRPWKIYSRTVYMQSYIGDFVQPSGWLEWN 1570
Query: 477 GNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGV 536
G LDT++YGE+ NYGPGS RV+WPG+ ++ D A FTV F G +WLP T +
Sbjct: 1571GTVGLDTIFYGEFNNYGPGSVTNNRVQWPGHFLLNDTQAWN-FTVLNFTLGNTWLPDTDI 1747
Query: 537 KFTAGLSN 544
+T GL N
Sbjct: 1748PYTEGLLN 1771
>TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylesterase isoform
alpha {Vigna radiata}, partial (94%)
Length = 1655
Score = 365 bits (938), Expect = e-101
Identities = 172/316 (54%), Positives = 232/316 (72%), Gaps = 9/316 (2%)
Frame = +2
Query: 221 WMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKE 280
W++ D++LL++ + D VVA DG+G + TI EA+ AAP+ S R VI++K G Y E
Sbjct: 41 WVSPKDRKLLQAAVNQTKFDLVVAKDGTGNFATIGEAIAAAPNSSATRFVIHIKAGAYFE 220
Query: 281 NIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAG 340
N+++ KK TN+M+VGDGIGQT+V ++RN + GWTTF+++TFAV G FIAK +TF N+AG
Sbjct: 221 NVEVIKKKTNLMLVGDGIGQTVVKASRNVVDGWTTFQSSTFAVVGDKFIAKGITFENSAG 400
Query: 341 PVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQ 400
P HQAVA+R +D SAF++CS QDTLY HSLRQFYREC++YGT+DFIFGN A V Q
Sbjct: 401 PSKHQAVAVRNGADFSAFYQCSFVAYQDTLYVHSLRQFYRECDVYGTVDFIFGNAAVVFQ 580
Query: 401 NCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLASQ---------PTYLGRPWK 451
NC +Y R P P QK TAQGR+ P+Q+TG +I + + A+ +YLGRPWK
Sbjct: 581 NCNLYARKPDPKQKNLFTAQGREDPNQNTGISILNCKIAAAADLIPVQSTFKSYLGRPWK 760
Query: 452 EYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIK 511
+YSRTVY+N+++ +++ P GWLEW G FALDTL+YGEY+N GPGS+ + RV WPGY VI
Sbjct: 761 KYSRTVYLNSFIDNLIDPPGWLEWNGTFALDTLFYGEYKNRGPGSNTSARVTWPGYKVIT 940
Query: 512 DASAAGYFTVQRFLNG 527
+A+ A FTV++F+ G
Sbjct: 941 NATEASQFTVRQFIQG 988
>TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1929
Score = 365 bits (937), Expect = e-101
Identities = 203/466 (43%), Positives = 279/466 (59%), Gaps = 29/466 (6%)
Frame = +3
Query: 106 AIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCIEGFEGT 165
A++DC+ L D + L+ S + + + Q E N++ LSA L+NQ TC++G + T
Sbjct: 315 ALQDCQSLSDLNFDFLSSSFQTVNKTTKFLPSLQGE-NIQTLLSAILTNQQTCLDGLKDT 491
Query: 166 DRR--LESYISGSVTQVTQLISNVLSLYTQ--LNRLPFRPPRNTTLHETSTDESLEFPEW 221
+ ++ ++ T+L S L+ +T+ +N + + H ++ P
Sbjct: 492 SSAWSFRNGLTIPLSNDTKLYSVSLAFFTKGWVNPKTNKTSFPNSKHSNKGFKNGRLPLK 671
Query: 222 MTEADQELLKSKPHGKIADA-------------VVALDGSGQYRTINEAVNAAPSHS--- 265
MT + + +S K+ + V+ DGSG + TIN+A+ AAP+ S
Sbjct: 672 MTSKTRAIYESVSRRKLLQSNQVGEDVVVRDIVTVSQDGSGNFTTINDAIAAAPNKSVSS 851
Query: 266 NRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSG 325
+ +IYV G+Y+E I + KK T +MM+GDGI +TI+T N + + GWTTF + TFAV G
Sbjct: 852 DGYFLIYVTAGVYEEYITIDKKKTYLMMIGDGINKTIITGNHSVVDGWTTFGSPTFAVVG 1031
Query: 326 KGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIY 385
+GF+ +MT RNTAG V HQAVALR +D S F+ CS EG QDTLY HSLRQFYREC+IY
Sbjct: 1032QGFVGVNMTIRNTAGAVKHQAVALRNGADLSTFYSCSFEGYQDTLYTHSLRQFYRECDIY 1211
Query: 386 GTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLASQ--- 442
GT+DFIFGN V QNC +Y R+P+ Q ITAQGR P+Q TG +I + + A+
Sbjct: 1212GTVDFIFGNAKVVFQNCNLYPRLPMSGQFNAITAQGRTDPNQDTGTSIHNCTIKATDDLA 1391
Query: 443 ------PTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGS 496
TYLGRPWKEYSRTVY+ T+M +++ GW W G FAL TL+Y E+ N GPGS
Sbjct: 1392ASNGAVSTYLGRPWKEYSRTVYMQTFMDNVINVAGWRAWDGEFALSTLYYAEFNNSGPGS 1571
Query: 497 SLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
S GRV W GYHVI +A+ A FTV FL G WLP+TGV +T L
Sbjct: 1572STDGRVTWQGYHVI-NATDAANFTVANFLLGDDWLPQTGVSYTNSL 1706
>NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1
Length = 1698
Score = 302 bits (774), Expect = 2e-82
Identities = 174/521 (33%), Positives = 280/521 (53%), Gaps = 16/521 (3%)
Frame = +1
Query: 37 QALIAQACMDIENQNSCLTNIHNELTRTGPPSPTSVINAALRTTINEAIGAINNMTKIST 96
Q + C + + +C H L + + + I AL T E INN
Sbjct: 157 QKNVDMLCQSTKFKETC----HKTLEKASFSNMKNRIKGALGATEEELRKHINNSALYQE 324
Query: 97 FSVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQD 156
+ ++ + A+E C E+LD++V + S+G + + + ++Y +++ WL+ LS+Q
Sbjct: 325 LATDSMTKQAMEICNEVLDYAVDGIHKSVGTLDQFDF-HKLSEYAFDIKVWLTGTLSHQQ 501
Query: 157 TCIEGFEGTDRRLESYISGSVTQVTQLISNVLSLYTQLNRL--PFRPPRNTTLHETSTDE 214
TC++GF T ++ + +L SN + + ++R+ F P + +D+
Sbjct: 502 TCLDGFVNTKTHAGETMAKVLKTSMELSSNAIDMMDVVSRILKGFHPSQYGVSRRLLSDD 681
Query: 215 SLEFPEWMTEADQELLKSKPHGKI-ADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYV 273
+ P W+++ + LL G + A+AVVA DGSGQ++T+ +A+ P + VIYV
Sbjct: 682 GI--PSWVSDGHRHLLAG---GNVKANAVVAQDGSGQFKTLTDALKTVPPTNAAPFVIYV 846
Query: 274 KKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDM 333
K G+YKE +++ K+M + ++GDG +T T + N+ G T++TATF V+G F+AKD+
Sbjct: 847 KAGVYKETVNVAKEMNYVTVIGDGPTKTKFTGSLNYADGINTYKTATFGVNGANFMAKDI 1026
Query: 334 TFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFG 393
F NTAG QAVALRV +DQ+ F C ++G QDTL+ S RQFYR+C I GTIDF+FG
Sbjct: 1027GFENTAGTSKFQAVALRVTADQAIFHNCQMDGFQDTLFVESQRQFYRDCAISGTIDFVFG 1206
Query: 394 NGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYV--------LASQPTY 445
+ V QNCK+ RVP QK +TA GR + ++ S+ + + +Y
Sbjct: 1207DAFGVFQNCKLICRVPAKGQKCLVTAGGRDKQNSASALVFLSSHFTGEPALTSVTPKLSY 1386
Query: 446 LGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWP 505
LGRPWK YS+ V +++ + +M P G++ +G DT + EY N GPG+ RVKW
Sbjct: 1387LGRPWKLYSKVVIMDSTIDAMFAPEGYMPMVGGAFKDTCTFYEYNNKGPGADTNLRVKWH 1566
Query: 506 GYHVIKDASAAGY-----FTVQRFLNGGSWLPRTGVKFTAG 541
G V+ AA Y F + +W+ ++GV ++ G
Sbjct: 1567GVKVLTSNVAAEYYPGKFFEIVNATARDTWIVKSGVPYSLG 1689
>TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectinesterase
{Arabidopsis thaliana}, partial (34%)
Length = 691
Score = 246 bits (627), Expect = 2e-65
Identities = 120/214 (56%), Positives = 157/214 (73%), Gaps = 1/214 (0%)
Frame = +3
Query: 217 EFPEWMTEADQELL-KSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKK 275
E P W+ D++LL K K AD VVA DGSG+Y+TI+ A+ P+ S++R VIYVKK
Sbjct: 48 EEPNWLHHKDRKLLQKDSDLKKKADIVVAKDGSGKYKTISAALKHVPNKSDKRTVIYVKK 227
Query: 276 GLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTF 335
G+Y EN+ ++K N+M++GDG+ TIV+ + NF+ G TF TATFAV G+ FIA+D+ F
Sbjct: 228 GIYYENVRVEKTKWNVMIIGDGMNVTIVSGSLNFIDGTPTFSTATFAVFGRNFIARDIGF 407
Query: 336 RNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNG 395
+NTAGP HQAVAL +DQ+ F++CS++ QDTLYAHS RQFYREC IYGT+DFIFGN
Sbjct: 408 KNTAGPQKHQAVALMTSADQAVFYKCSMDAYQDTLYAHSNRQFYRECNIYGTVDFIFGNS 587
Query: 396 AAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQST 429
A VLQNC I R P+P Q+ TITAQG+ P+ +T
Sbjct: 588 AVVLQNCNILPRQPMPGQQNTITAQGKTDPNMNT 689
>BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precursor (EC
3.1.1.11) (Pectin methylesterase) (PE). [Peach] {Prunus
persica}, partial (36%)
Length = 589
Score = 212 bits (539), Expect(2) = 2e-59
Identities = 97/161 (60%), Positives = 130/161 (80%)
Frame = +2
Query: 247 GSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSN 306
GSG Y + +AV+AAP S +R+VIYVKKG+Y EN+++KKK NIM++G+G+ TI++ +
Sbjct: 2 GSGDYAKVMDAVSAAPESSMKRYVIYVKKGVYVENVEIKKKKWNIMLIGEGMDATIISGS 181
Query: 307 RNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGN 366
RN++ G TTFR+ATFAVSG+GFIA+D++F+NTAG HQAVALR DSD S F+RC I G
Sbjct: 182 RNYVDGSTTFRSATFAVSGRGFIARDISFQNTAGAEKHQAVALRSDSDLSVFYRCGIFGY 361
Query: 367 QDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTR 407
QD+LY H++RQFYREC+I GT+DFIFG+ AV QNC+I +
Sbjct: 362 QDSLYTHTMRQFYRECKISGTVDFIFGDATAVFQNCQILAK 484
Score = 36.2 bits (82), Expect(2) = 2e-59
Identities = 15/23 (65%), Positives = 19/23 (82%)
Frame = +3
Query: 410 LPLQKVTITAQGRKSPHQSTGFT 432
+P QK T+TAQGRK P+Q TGF+
Sbjct: 492 MPKQKNTVTAQGRKDPNQPTGFS 560
>BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like protein -
Arabidopsis thaliana, partial (36%)
Length = 658
Score = 205 bits (521), Expect = 4e-53
Identities = 108/218 (49%), Positives = 138/218 (62%), Gaps = 20/218 (9%)
Frame = +1
Query: 256 EAVNAAPSHS--NRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQ-G 312
EAVNAAP + +R VIY+K+G+Y+E + + K N++ +GDGIG+T++T + N Q G
Sbjct: 4 EAVNAAPDNGVDRKRFVIYIKEGVYEETVRVPLKKRNVVFLGDGIGKTVITGSANVGQPG 183
Query: 313 WTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYA 372
TT+ +AT AV G GF+AKD+T NTAGP HQAVA R+DSD S C GNQDTLYA
Sbjct: 184 MTTYNSATVAVLGDGFMAKDLTIENTAGPDAHQAVAFRLDSDLSVIENCEFLGNQDTLYA 363
Query: 373 HSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTR----VPLPLQKVTITAQGRKSPHQS 428
HSLRQFY+ C I G +DFIFGN AA+ Q+C+I R P + ITA GR P QS
Sbjct: 364 HSLRQFYKSCRIVGNVDFIFGNSAAIFQDCQILVRPRQLKPEKGENNAITAHGRTDPAQS 543
Query: 429 TGFTIQDSYVLASQ-------------PTYLGRPWKEY 453
TGF Q+ + ++ YLGRPWKEY
Sbjct: 544 TGFVFQNCLINGTEDYMALYHSNPKVHKNYLGRPWKEY 657
>BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-like protein
- maize, partial (25%)
Length = 678
Score = 179 bits (454), Expect = 2e-45
Identities = 101/218 (46%), Positives = 124/218 (56%), Gaps = 14/218 (6%)
Frame = +1
Query: 268 RHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKG 327
R+ IYVK G+Y E I + K NI+M GDG G+TIVT +N G T +TATFA + G
Sbjct: 4 RYTIYVKAGVYDEYITIPKDAVNILMYGDGPGKTIVTGRKNGAAGVKTMQTATFANTALG 183
Query: 328 FIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGT 387
FI K MTF NTAGP HQAVA R D SA C I G QDTLY + RQFYR C I GT
Sbjct: 184 FIGKAMTFENTAGPAGHQAVAFRNQGDMSALVGCHILGYQDTLYVQTNRQFYRNCVISGT 363
Query: 388 IDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLASQ----- 442
+DFIFG A ++Q+ I R+P P Q TITA G +TG IQ ++
Sbjct: 364 VDFIFGTSATLIQDSTIIVRMPSPNQFNTITADGSYVNKLNTGIVIQGCNIVPKAALFPQ 543
Query: 443 ----PTYLGRPWKEYSRTVYINT-----YMSSMVQPRG 471
+YLGRPWK ++TV + + Y S + P G
Sbjct: 544 RFTIKSYLGRPWKVLTKTVVMESTIG*FYSSXWMDPLG 657
>TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pectin
methylesterase {Arabidopsis thaliana}, partial (33%)
Length = 1289
Score = 137 bits (346), Expect(2) = 5e-45
Identities = 79/220 (35%), Positives = 123/220 (55%), Gaps = 12/220 (5%)
Frame = +3
Query: 243 VALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTI 302
V+ DGS Q+++I EA+N+ ++ RR +I + G Y+E I + K + I +GD
Sbjct: 243 VSQDGSAQFKSITEALNSIQPYNIRRVIISIAPGYYREKIVVPKTLPFITFLGDVRDPPT 422
Query: 303 VTSNRNFMQGWT--------TFRTATFAVSGKGFIAKDMTFRNTA----GPVNHQAVALR 350
+T N Q T TF +AT AV+ F+A ++ F NTA G QAVA+R
Sbjct: 423 ITGNDT--QSVTGSDGAQLRTFNSATVAVNASYFMAININFENTASFPIGSKVEQAVAVR 596
Query: 351 VDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPL 410
+ +++AF+ C+ G QDTLY H ++ C I G++DFI G+G ++ + C I + +
Sbjct: 597 ITGNKTAFYNCTFSGVQDTLYDHKGLHYFNNCTIKGSVDFICGHGKSLYEGCTIRS---I 767
Query: 411 PLQKVTITAQGRKSPHQSTGFTIQDSYVLASQPTYLGRPW 450
+ITAQ +P +GF+ ++S V+ PTYLGRPW
Sbjct: 768 ANNMTSITAQSGSNPSYDSGFSFKNSMVIGDGPTYLGRPW 887
Score = 62.0 bits (149), Expect(2) = 5e-45
Identities = 30/79 (37%), Positives = 45/79 (55%)
Frame = +1
Query: 453 YSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKD 512
YS+ V+ TYM + V P+GW +W +YGEY+ GPGS+ AGRV W ++ D
Sbjct: 895 YSQVVFSYTYMDNSVLPKGWEDWNDTKRYMNAYYGEYKCSGPGSNTAGRVPWA--RMLND 1068
Query: 513 ASAAGYFTVQRFLNGGSWL 531
A + Q +++G +WL
Sbjct: 1069KEAQVFIGTQ-YIDGNTWL 1122
>TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor (EC
3.1.1.11) (Pectin methylesterase) (PE) (P65). [Alfalfa],
partial (53%)
Length = 732
Score = 173 bits (438), Expect = 2e-43
Identities = 84/200 (42%), Positives = 118/200 (59%), Gaps = 8/200 (4%)
Frame = +2
Query: 328 FIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGT 387
F A ++ F N+AG HQAVALRV +D++ F+ C + G QDTLY S RQFYR+C I GT
Sbjct: 86 FTALNVGFENSAGAAKHQAVALRVTADKALFYNCQMNGYQDTLYTQSKRQFYRDCTITGT 265
Query: 388 IDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSY------VLAS 441
IDF+FG+ V QNCK+ R P+ Q+ +TA GR + Q+ + VL
Sbjct: 266 IDFVFGDAVGVFQNCKLIVRKPMANQQCMVTAGGRTKVDSVSALVFQNCHFTGEPEVLTM 445
Query: 442 QP--TYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLA 499
QP YLGRPW+ +S+ V +++ + + P G++ W+GN +T Y EY N G G++
Sbjct: 446 QPKIAYLGRPWRNFSKVVIVDSLIDGLFVPEGYMPWMGNLFKETCTYLEYNNKGAGAATN 625
Query: 500 GRVKWPGYHVIKDASAAGYF 519
+VKWPG I AA Y+
Sbjct: 626 LKVKWPGVKTISAGEAAKYY 685
>AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (25%)
Length = 495
Score = 169 bits (427), Expect = 3e-42
Identities = 80/144 (55%), Positives = 108/144 (74%), Gaps = 3/144 (2%)
Frame = +1
Query: 246 DGSGQYRTINEAVNAAPSH---SNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTI 302
DGSG + TIN+A+ AAP++ S+ +I++ +G+Y+E + + KK +MM+G+GI QTI
Sbjct: 64 DGSGNFTTINDAIAAAPNNTVASDGYFLIFITEGVYEEYVSIDKKKKYLMMIGEGINQTI 243
Query: 303 VTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCS 362
+T NRN G+TTF +ATFAV +GF+A ++TFRNTAG +QAVALR +D S F+ CS
Sbjct: 244 ITGNRNVADGFTTFNSATFAVVAQGFVAVNITFRNTAGAAKNQAVALRSGADMSTFYSCS 423
Query: 363 IEGNQDTLYAHSLRQFYRECEIYG 386
EG QDTLY HSLRQFYREC+IYG
Sbjct: 424 FEGYQDTLYTHSLRQFYRECDIYG 495
>TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE8B
precursor (EC 3.1.1.11) (Pectin methylesterase) (PE).
[Peach] {Prunus persica}, partial (34%)
Length = 1415
Score = 159 bits (402), Expect = 3e-39
Identities = 93/244 (38%), Positives = 144/244 (58%), Gaps = 1/244 (0%)
Frame = +1
Query: 79 TTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDR-T 137
T + + I ++ T + S R A+ DC +LLD S+ +L S+ ++ + D T
Sbjct: 253 TVLQKVISILSRFTNVFGHS---RTSNAVSDCLDLLDMSLDQLNQSISAAQKPKEKDNST 423
Query: 138 AQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQVTQLISNVLSLYTQLNRL 197
+ +L WLSA L DTCIEG EG+ ++ IS + V L++N+L N
Sbjct: 424 GKLNCDLRTWLSAVLVYPDTCIEGLEGSI--VKGLISSGLDHVMSLVANLLGEVVSGND- 594
Query: 198 PFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEA 257
+ +T++ FP W+ + D +LL++ +G ADAVVA DGSG Y + +A
Sbjct: 595 ----------DQLATNKD-RFPSWIRDEDTKLLQA--NGVTADAVVAADGSGDYAKVMDA 735
Query: 258 VNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFR 317
V+AAP S +R+VIYVKKG+Y EN+++KKK NIM++G+G+ TI++ +RN++ G TTFR
Sbjct: 736 VSAAPESSMKRYVIYVKKGVYVENVEIKKKKWNIMLIGEGMDATIISGSRNYVDGSTTFR 915
Query: 318 TATF 321
+ATF
Sbjct: 916 SATF 927
>TC93392 similar to PIR|T49241|T49241 pectinesterase-like protein -
Arabidopsis thaliana, partial (18%)
Length = 467
Score = 138 bits (347), Expect = 6e-33
Identities = 59/98 (60%), Positives = 75/98 (76%)
Frame = +2
Query: 445 YLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKW 504
YLGRPWK YSRT+++ +YMS ++P GWLEW GNFAL+TL+Y EY N GPG+ +A RVKW
Sbjct: 2 YLGRPWKTYSRTIFMQSYMSDAIRPEGWLEWNGNFALNTLYYAEYMNSGPGAGVANRVKW 181
Query: 505 PGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
GYHV+ D+S A FTV +F+ G LP TGV +T+GL
Sbjct: 182 SGYHVLNDSSEATKFTVAQFIEGNLGLPSTGVTYTSGL 295
>TC82059 similar to GP|19424045|gb|AAL87311.1 unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 483
Score = 135 bits (339), Expect = 5e-32
Identities = 61/104 (58%), Positives = 76/104 (72%)
Frame = +1
Query: 439 LASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSL 498
+A+ TYLGRPWKEYSRTVY+ ++M S + P GW EW G+FAL TL+Y EY N G GSS
Sbjct: 4 VATVKTYLGRPWKEYSRTVYMQSFMDSFINPSGWREWNGDFALSTLYYAEYDNRGAGSST 183
Query: 499 AGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
A RV WPGYHVI A+ A FTV F++G W+P+TGV + +GL
Sbjct: 184 ANRVTWPGYHVI-GATDAANFTVSNFVSGDDWIPQTGVPYMSGL 312
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,101,684
Number of Sequences: 36976
Number of extensions: 216293
Number of successful extensions: 1154
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 1087
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1095
length of query: 544
length of database: 9,014,727
effective HSP length: 101
effective length of query: 443
effective length of database: 5,280,151
effective search space: 2339106893
effective search space used: 2339106893
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144513.13


