

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.7 + phase: 0
(284 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
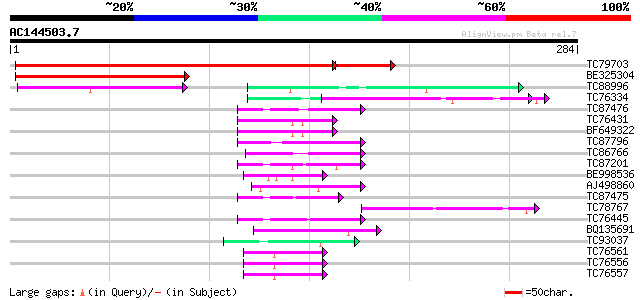
Score E
Sequences producing significant alignments: (bits) Value
TC79703 homologue to GP|21615411|emb|CAD33925. proline rich prot... 342 e-107
BE325304 homologue to GP|21615411|emb proline rich protein 3 {Ci... 177 5e-45
TC88996 similar to PIR|T00933|T00933 RNA-binding protein homolog... 73 1e-13
TC76334 similar to GP|9663767|emb|CAC01237.1 RNA Binding Protein... 46 1e-05
TC87476 homologue to PIR|T11622|T11622 extensin class 1 precurso... 45 4e-05
TC76431 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sat... 44 7e-05
BF649322 similar to GP|15021754|gb| root nodule extensin {Pisum ... 44 7e-05
TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear prot... 44 7e-05
TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E... 44 9e-05
TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG292... 44 9e-05
BE998536 similar to GP|2598593|emb| MtN12 {Medicago truncatula},... 42 2e-04
AJ498860 similar to GP|20197423|gb|A unknown protein {Arabidopsi... 42 3e-04
TC87475 similar to PIR|T11622|T11622 extensin class 1 precursor ... 42 3e-04
TC78767 similar to PIR|S59117|S59117 small nuclear ribonucleopro... 42 3e-04
TC76445 similar to PIR|T07623|T07623 extensin homolog HRGP2 - so... 42 3e-04
BQ135691 similar to PIR|E96636|E966 hypothetical protein T7P1.21... 42 3e-04
TC93037 weakly similar to GP|3004934|gb|AAC09237.1| annexin XIV ... 41 5e-04
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 41 5e-04
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 41 5e-04
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 41 5e-04
>TC79703 homologue to GP|21615411|emb|CAD33925. proline rich protein 3
{Cicer arietinum}, partial (67%)
Length = 812
Score = 342 bits (877), Expect(2) = e-107
Identities = 160/161 (99%), Positives = 161/161 (99%)
Frame = +1
Query: 4 QVRTIFITGLPEDVKERELQNLCRWLPGFEASQLNFKAEKPMGFALFNSPHQAIAAKDIL 63
+VRTIFITGLPEDVKERELQNLCRWLPGFEASQLNFKAEKPMGFALFNSPHQAIAAKDIL
Sbjct: 241 EVRTIFITGLPEDVKERELQNLCRWLPGFEASQLNFKAEKPMGFALFNSPHQAIAAKDIL 420
Query: 64 QDMLFDPEAKSVLHTEMAKKNLFVKRGIGADAVAFDQSKRLRTAGDYNHTGYVTPSPFHP 123
QDMLFDPEAKSVLHTEMAKKNLFVKRGIGADAVAFDQSKRLRTAGDYNHTGYVTPSPFHP
Sbjct: 421 QDMLFDPEAKSVLHTEMAKKNLFVKRGIGADAVAFDQSKRLRTAGDYNHTGYVTPSPFHP 600
Query: 124 PPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAP 164
PPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAP
Sbjct: 601 PPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAP 723
Score = 65.5 bits (158), Expect(2) = e-107
Identities = 29/30 (96%), Positives = 29/30 (96%)
Frame = +2
Query: 164 PSSYVPIQNTKDNPPCNTLFIGNLGENINE 193
PSSYVPIQNTKDNP CNTLFIGNLGENINE
Sbjct: 722 PSSYVPIQNTKDNPSCNTLFIGNLGENINE 811
Score = 36.2 bits (82), Expect = 0.015
Identities = 37/152 (24%), Positives = 56/152 (36%), Gaps = 1/152 (0%)
Frame = +1
Query: 120 PFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNPPC 179
P+H PP APPPP P +P P+ +
Sbjct: 163 PYHQQWPPA------AAPPPPQP---------TAIPSPSSEEV----------------- 246
Query: 180 NTLFIGNLGENINEEEVRGLFSVQPGFKQMKIIRQERHTVCFIEFEDVNSATNVHHNLQG 239
T+FI L E++ E E++ L PGF+ ++ + + F F + A LQ
Sbjct: 247 RTIFITGLPEDVKERELQNLCRWLPGFEASQLNFKAEKPMGFALFNSPHQAIAAKDILQD 426
Query: 240 AVI-PSSGSIGMRIQYSKNPFGKRKDGTYPMA 270
+ P + S+ KN F KR G +A
Sbjct: 427 MLFDPEAKSVLHTEMAKKNLFVKRGIGADAVA 522
>BE325304 homologue to GP|21615411|emb proline rich protein 3 {Cicer
arietinum}, partial (31%)
Length = 447
Score = 177 bits (448), Expect = 5e-45
Identities = 86/87 (98%), Positives = 87/87 (99%)
Frame = +3
Query: 4 QVRTIFITGLPEDVKERELQNLCRWLPGFEASQLNFKAEKPMGFALFNSPHQAIAAKDIL 63
+VRTIFITGLPEDVKERELQNLCRWLPGFEASQLNFKAEKPMGFALFNSPHQAIAAKDIL
Sbjct: 12 EVRTIFITGLPEDVKERELQNLCRWLPGFEASQLNFKAEKPMGFALFNSPHQAIAAKDIL 191
Query: 64 QDMLFDPEAKSVLHTEMAKKNLFVKRG 90
QDMLFDPEAKSVLHTEMAKKNLFVKRG
Sbjct: 192QDMLFDPEAKSVLHTEMAKKNLFVKRG 272
Score = 30.0 bits (66), Expect = 1.1
Identities = 23/83 (27%), Positives = 37/83 (43%), Gaps = 1/83 (1%)
Frame = +3
Query: 181 TLFIGNLGENINEEEVRGLFSVQPGFKQMKIIRQERHTVCFIEFEDVNSATNVHHNLQGA 240
T+FI L E++ E E++ L PGF+ ++ + + F F + A LQ
Sbjct: 21 TIFITGLPEDVKERELQNLCRWLPGFEASQLNFKAEKPMGFALFNSPHQAIAAKDILQDM 200
Query: 241 VI-PSSGSIGMRIQYSKNPFGKR 262
+ P + S+ KN F KR
Sbjct: 201 LFDPEAKSVLHTEMAKKNLFVKR 269
>TC88996 similar to PIR|T00933|T00933 RNA-binding protein homolog At2g42240
- Arabidopsis thaliana, partial (51%)
Length = 651
Score = 73.2 bits (178), Expect = 1e-13
Identities = 40/87 (45%), Positives = 53/87 (59%), Gaps = 2/87 (2%)
Frame = +2
Query: 5 VRTIFITGLPEDVKERELQNLCRWLPGFEASQLNF--KAEKPMGFALFNSPHQAIAAKDI 62
VRT+FI GLPEDVK RE+ NL R PG+E+S L + + FA+F++ AI A
Sbjct: 269 VRTLFIAGLPEDVKPREIYNLFREFPGYESSHLRSPNNSSQAFAFAVFSNQQSAIMALHA 448
Query: 63 LQDMLFDPEAKSVLHTEMAKKNLFVKR 89
L M+FD E S L+ ++AK N KR
Sbjct: 449 LNGMIFDLEKGSTLYIDLAKSNSRAKR 529
Score = 54.3 bits (129), Expect = 5e-08
Identities = 40/144 (27%), Positives = 58/144 (39%), Gaps = 6/144 (4%)
Frame = +2
Query: 120 PFHPPPPPVWGPHGYMAPPP----PPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKD 175
P PPPPP + P + PPP PPP+ P+ S PS P+ +T D
Sbjct: 98 PQQPPPPPQYQPIPVIPPPPGAPLPPPHHPHPHPQYIPQQQQVYDSYVPS---PLASTND 268
Query: 176 NPPCNTLFIGNLGENINEEEVRGLFSVQPGFK--QMKIIRQERHTVCFIEFEDVNSATNV 233
TLFI L E++ E+ LF PG++ ++ F F + SA
Sbjct: 269 ---VRTLFIAGLPEDVKPREIYNLFREFPGYESSHLRSPNNSSQAFAFAVFSNQQSAIMA 439
Query: 234 HHNLQGAVIPSSGSIGMRIQYSKN 257
H L G + + I +K+
Sbjct: 440 LHALNGMIFDLEKGSTLYIDLAKS 511
>TC76334 similar to GP|9663767|emb|CAC01237.1 RNA Binding Protein 45
{Nicotiana plumbaginifolia}, partial (68%)
Length = 1579
Score = 46.2 bits (108), Expect = 1e-05
Identities = 39/147 (26%), Positives = 57/147 (38%), Gaps = 4/147 (2%)
Frame = +1
Query: 120 PFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNPPC 179
P H PP G PPP PY V P P A S P Q
Sbjct: 61 PGHGMAPPTMGQQPPQQYQQPPPQQPY----VMMQPQQPPAMWAAQSAQPPQQPASADEV 228
Query: 180 NTLFIGNLGENINEEEVRGLFSVQPGFKQMKIIRQERHTVC----FIEFEDVNSATNVHH 235
TL+IG+L ++E + F +K+IR ++ + FIEF +A V
Sbjct: 229 RTLWIGDLQYWMDENYLYTCFGNTGELTSVKVIRNKQTSQSEGYGFIEFNTRATAERVLQ 408
Query: 236 NLQGAVIPSSGSIGMRIQYSKNPFGKR 262
G ++P+ G R+ ++ G+R
Sbjct: 409 TYNGTIMPNGGQ-NYRLNWATFSAGER 486
Score = 45.8 bits (107), Expect = 2e-05
Identities = 33/120 (27%), Positives = 56/120 (46%), Gaps = 6/120 (5%)
Frame = +1
Query: 157 PAPVSIAPSSYVPIQNTKDNPPCNT-LFIGNLGENINEEEVRGLFSVQPGFKQMKIIRQE 215
PA + +SY P +N P NT +F+GNL N+ ++ +R +F+ +KI +
Sbjct: 760 PAATTQPKASYNPSGGQSENDPNNTTIFVGNLDPNVTDDHLRQVFTQYGELVHVKIPSGK 939
Query: 216 RHTVCFIEFEDVNSATNVHHNLQGAVIPSSGSIGMRIQYSKNPFGKR-----KDGTYPMA 270
R F++F D + A L G ++ G +R+ + + P K+ G YP A
Sbjct: 940 R--CGFVQFSDRSCAEEAIRVLNGTLL---GGQNVRLSWGRTPSNKQTQQDPTQGGYPAA 1104
Score = 26.9 bits (58), Expect = 8.9
Identities = 17/54 (31%), Positives = 22/54 (40%), Gaps = 3/54 (5%)
Frame = +1
Query: 105 RTAGDYNHTGYVTPSPFHPPPPPVWGPHGYMAPP---PPPPYDPYAGYPVAQVP 155
+T D GY ++ + +GY APP P Y Y GYP Q P
Sbjct: 1063 QTQQDPTQGGYPAAGGYYGYAQGGYENYGYAAPPAGQDPNVYGSYPGYPGYQHP 1224
>TC87476 homologue to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (48%)
Length = 939
Score = 44.7 bits (104), Expect = 4e-05
Identities = 25/64 (39%), Positives = 29/64 (45%)
Frame = +3
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
Y +P P P PPP P+GY +PPPP P P P P P S +P Y P
Sbjct: 546 YKSPPPPSPSPPP---PYGYKSPPPPSPSPP----PPYIYKSPPPPSPSPPPYHPYLYNS 704
Query: 175 DNPP 178
PP
Sbjct: 705 PPPP 716
Score = 39.3 bits (90), Expect = 0.002
Identities = 22/64 (34%), Positives = 31/64 (48%), Gaps = 5/64 (7%)
Frame = +2
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPP-----PPPPYDPYAGYPVAQVPMPAPVSIAPSSYVP 169
Y +P P P PPP P+ Y +PP PPPPY + P + P P + +P + +P
Sbjct: 209 YKSPPPPSPSPPP---PYVYKSPPPPSPSPPPPYVYKSPPPPSPSPPPPYIYKSPPTTIP 379
Query: 170 IQNT 173
T
Sbjct: 380 FTTT 391
Score = 37.0 bits (84), Expect = 0.009
Identities = 21/68 (30%), Positives = 29/68 (41%)
Frame = +3
Query: 111 NHTGYVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPI 170
+H +++ S PP P P+ Y +PPPP P P Y P P+P P Y
Sbjct: 333 HHRLHISTSHHRPPSPSPPPPYVYKSPPPPSP-SPPPPYVYKSPPPPSPSPPPPYVYKSP 509
Query: 171 QNTKDNPP 178
+PP
Sbjct: 510 PPPSASPP 533
Score = 36.6 bits (83), Expect = 0.011
Identities = 22/64 (34%), Positives = 27/64 (41%)
Frame = +3
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
Y +P P P PPP P+ Y +PPP P P Y P P+P P Y
Sbjct: 450 YKSPPPPSPSPPP---PYVYKSPPP-PSASPPPPYYYKSPPPPSPSPPPPYGYKSPPPPS 617
Query: 175 DNPP 178
+PP
Sbjct: 618 PSPP 629
Score = 35.8 bits (81), Expect = 0.019
Identities = 22/60 (36%), Positives = 28/60 (46%), Gaps = 5/60 (8%)
Frame = +2
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPP-----PPPPYDPYAGYPVAQVPMPAPVSIAPSSYVP 169
Y +P P P PPP P+ Y +PP PPPPY + P + P P V +P P
Sbjct: 17 YKSPPPPSPSPPP---PYVYKSPPPPSPSPPPPYVYKSPPPPSPSPPPPYVYKSPPPPTP 187
Score = 35.4 bits (80), Expect = 0.025
Identities = 22/64 (34%), Positives = 26/64 (40%)
Frame = +2
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
Y +P P P PPP P+ Y +PPP P P Y P P P P Y
Sbjct: 65 YKSPPPPSPSPPP---PYVYKSPPP-PSPSPPPPYVYKSPPPPTPSPPPPYIYKSPPPPS 232
Query: 175 DNPP 178
+PP
Sbjct: 233 PSPP 244
Score = 35.4 bits (80), Expect = 0.025
Identities = 22/64 (34%), Positives = 27/64 (41%)
Frame = +2
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
Y +P P P PPP P+ Y +PPP P P Y P P+P P Y
Sbjct: 113 YKSPPPPSPSPPP---PYVYKSPPP-PTPSPPPPYIYKSPPPPSPSPPPPYVYKSPPPPS 280
Query: 175 DNPP 178
+PP
Sbjct: 281 PSPP 292
Score = 35.0 bits (79), Expect = 0.033
Identities = 22/64 (34%), Positives = 27/64 (41%)
Frame = +2
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
Y +P P P PPP P+ Y +PPP P P Y P P+P P Y
Sbjct: 161 YKSPPPPTPSPPP---PYIYKSPPP-PSPSPPPPYVYKSPPPPSPSPPPPYVYKSPPPPS 328
Query: 175 DNPP 178
+PP
Sbjct: 329 PSPP 340
Score = 34.3 bits (77), Expect = 0.056
Identities = 21/64 (32%), Positives = 26/64 (39%), Gaps = 3/64 (4%)
Frame = +3
Query: 118 PSPFHPPPPPVWG---PHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
P + PPPP P+ Y +PPPP P P Y P P+P P Y
Sbjct: 489 PYVYKSPPPPSASPPPPYYYKSPPPPSP-SPPPPYGYKSPPPPSPSPPPPYIYKSPPPPS 665
Query: 175 DNPP 178
+PP
Sbjct: 666 PSPP 677
Score = 33.9 bits (76), Expect = 0.073
Identities = 20/55 (36%), Positives = 26/55 (46%)
Frame = +3
Query: 114 GYVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYV 168
GY +P P P PPP P+ Y +PPP P P +P P P+ A Y+
Sbjct: 591 GYKSPPPPSPSPPP---PYIYKSPPP-PSPSPPPYHPYLYNSPPPPIY*ASGYYL 743
Score = 33.1 bits (74), Expect = 0.12
Identities = 21/64 (32%), Positives = 26/64 (39%), Gaps = 3/64 (4%)
Frame = +2
Query: 118 PSPFHPPPPPVWG---PHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
P + PPPP P+ Y +PPPP P P Y P P+P P Y
Sbjct: 8 PYIYKSPPPPSPSPPPPYVYKSPPPPSP-SPPPPYVYKSPPPPSPSPPPPYVYKSPPPPT 184
Query: 175 DNPP 178
+PP
Sbjct: 185 PSPP 196
Score = 32.7 bits (73), Expect = 0.16
Identities = 18/41 (43%), Positives = 21/41 (50%), Gaps = 4/41 (9%)
Frame = +3
Query: 110 YNHTGYVTPSPFHPPPPPVWGPHGYMAPPP----PPPYDPY 146
Y + PSP PPPP + Y +PPP PPPY PY
Sbjct: 588 YGYKSPPPPSP-SPPPPYI-----YKSPPPPSPSPPPYHPY 692
Score = 31.2 bits (69), Expect = 0.47
Identities = 22/64 (34%), Positives = 26/64 (40%), Gaps = 3/64 (4%)
Frame = +3
Query: 118 PSPFHPPPPPVWG---PHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
P + PPPP P+ Y +PPPP P P P P P S +P P K
Sbjct: 393 PYVYKSPPPPSPSPPPPYVYKSPPPPSPSPP----PPYVYKSPPPPSASPP---PPYYYK 551
Query: 175 DNPP 178
PP
Sbjct: 552 SPPP 563
>TC76431 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sativum},
partial (30%)
Length = 890
Score = 43.9 bits (102), Expect = 7e-05
Identities = 23/55 (41%), Positives = 30/55 (53%), Gaps = 5/55 (9%)
Frame = +1
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPP--PPYDP---YAGYPVAQVPMPAPVSIAP 164
+V+P P++ PPP P+ Y +PPPP PPY P Y PV P PV +P
Sbjct: 244 HVSPKPYYHSPPPPKKPYKYSSPPPPVHPPYTPKPVYHSPPVHPPYTPHPVYHSP 408
Score = 35.8 bits (81), Expect = 0.019
Identities = 16/28 (57%), Positives = 18/28 (64%), Gaps = 1/28 (3%)
Frame = +1
Query: 117 TPSP-FHPPPPPVWGPHGYMAPPPPPPY 143
TP P +H PPPPV H Y+ PPPPY
Sbjct: 382 TPHPVYHSPPPPV---HSYIYASPPPPY 456
Score = 33.1 bits (74), Expect = 0.12
Identities = 22/59 (37%), Positives = 25/59 (42%), Gaps = 12/59 (20%)
Frame = +1
Query: 118 PSPFHPPPPPVWGPHGYMAP------PPPPPYDPYAGYPVA------QVPMPAPVSIAP 164
P P+HP P P PH + P PPP P PY YP P P PV +P
Sbjct: 25 PPPYHPYPHPHPHPHPHPHPYPVHHSPPPSPKKPYK-YPSPPPPTHHHYPHPHPVYHSP 198
Score = 32.7 bits (73), Expect = 0.16
Identities = 20/63 (31%), Positives = 27/63 (42%), Gaps = 3/63 (4%)
Frame = +1
Query: 118 PSPF---HPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
P P+ H PPP P+ Y +PPPP + +PV P P P S P+ +
Sbjct: 76 PHPYPVHHSPPPSPKKPYKYPSPPPPTHHHYPHPHPVYHSP-PPPKYKYSSPPPPVHHVS 252
Query: 175 DNP 177
P
Sbjct: 253 PKP 261
Score = 32.3 bits (72), Expect = 0.21
Identities = 24/67 (35%), Positives = 29/67 (42%), Gaps = 9/67 (13%)
Frame = +1
Query: 112 HTGYVTPSP----FHPPPPPVW--GPHGYMAPPPPP--PYDPYAGYPVAQVP-MPAPVSI 162
H Y +P P + PPPPV P Y PPPP PY + P P P PV
Sbjct: 178 HPVYHSPPPPKYKYSSPPPPVHHVSPKPYYHSPPPPKKPYKYSSPPPPVHPPYTPKPVYH 357
Query: 163 APSSYVP 169
+P + P
Sbjct: 358 SPPVHPP 378
Score = 29.3 bits (64), Expect = 1.8
Identities = 16/42 (38%), Positives = 21/42 (49%)
Frame = +1
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
P + PPPP + P+ + P P P P YPV P P+P
Sbjct: 4 PYKYSSPPPP-YHPYPHPHPHPHPHPHP---YPVHHSPPPSP 117
Score = 28.1 bits (61), Expect = 4.0
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 3/37 (8%)
Frame = +1
Query: 117 TPSPFHPPPP--PVWGPHG-YMAPPPPPPYDPYAGYP 150
TP P + PP P + PH Y +PPPP YA P
Sbjct: 337 TPKPVYHSPPVHPPYTPHPVYHSPPPPVHSYIYASPP 447
Score = 27.7 bits (60), Expect = 5.2
Identities = 19/56 (33%), Positives = 21/56 (36%), Gaps = 9/56 (16%)
Frame = +1
Query: 118 PSPFHP-----PPPPVWG----PHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAP 164
PSP P PPPP PH PPPP Y + P P P +P
Sbjct: 109 PSPKKPYKYPSPPPPTHHHYPHPHPVYHSPPPPKYKYSSPPPPVHHVSPKPYYHSP 276
>BF649322 similar to GP|15021754|gb| root nodule extensin {Pisum sativum},
partial (54%)
Length = 476
Score = 43.9 bits (102), Expect = 7e-05
Identities = 23/55 (41%), Positives = 30/55 (53%), Gaps = 5/55 (9%)
Frame = +1
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPP--PPYDP---YAGYPVAQVPMPAPVSIAP 164
+V+P P++ PPP P+ Y +PPPP PPY P Y PV P PV +P
Sbjct: 265 HVSPKPYYHSPPPPKKPYKYSSPPPPVHPPYTPKPVYHSPPVHPPYTPHPVYHSP 429
Score = 32.3 bits (72), Expect = 0.21
Identities = 24/67 (35%), Positives = 29/67 (42%), Gaps = 9/67 (13%)
Frame = +1
Query: 112 HTGYVTPSP----FHPPPPPVW--GPHGYMAPPPPP--PYDPYAGYPVAQVP-MPAPVSI 162
H Y +P P + PPPPV P Y PPPP PY + P P P PV
Sbjct: 199 HPVYHSPPPPKYKYSSPPPPVHHVSPKPYYHSPPPPKKPYKYSSPPPPVHPPYTPKPVYH 378
Query: 163 APSSYVP 169
+P + P
Sbjct: 379 SPPVHPP 399
Score = 29.3 bits (64), Expect = 1.8
Identities = 14/27 (51%), Positives = 16/27 (58%), Gaps = 1/27 (3%)
Frame = +1
Query: 117 TPSP-FHPPPPPVWGPHGYMAPPPPPP 142
TP P +H PPPPV H Y+ PP P
Sbjct: 403 TPHPVYHSPPPPV---HSYIYAXPPXP 474
>TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear protein.
[strain B95-8 Human herpesvirus 4] {Epstein-barr
virus}, partial (23%)
Length = 431
Score = 43.9 bits (102), Expect = 7e-05
Identities = 25/64 (39%), Positives = 27/64 (42%)
Frame = -1
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
Y P PPPPP P PPPPPP DP P P+P P AP + P
Sbjct: 203 YAPPCAPPPPPPPPEPP-----PPPPPPGDPPPYPPPVPPPLPYPTPCAPPAPPPPPPPL 39
Query: 175 DNPP 178
PP
Sbjct: 38 PPPP 27
Score = 35.8 bits (81), Expect = 0.019
Identities = 22/53 (41%), Positives = 23/53 (42%), Gaps = 1/53 (1%)
Frame = -1
Query: 118 PSPFHPPPPPVWG-PHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVP 169
P P PPPPP G P Y PPP PP PY P P P + P P
Sbjct: 170 PPPEPPPPPPPPGDPPPY--PPPVPPPLPYPTPCAPPAPPPPPPPLPPPPPYP 18
Score = 30.4 bits (67), Expect = 0.81
Identities = 18/43 (41%), Positives = 18/43 (41%), Gaps = 15/43 (34%)
Frame = -1
Query: 118 PSPFHPPP--PPVWGPHGYMAP-------------PPPPPYDP 145
P P PPP PPV P Y P PPPPPY P
Sbjct: 143 PPPGDPPPYPPPVPPPLPYPTPCAPPAPPPPPPPLPPPPPYPP 15
>TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E8.80 -
Arabidopsis thaliana, partial (18%)
Length = 1260
Score = 43.5 bits (101), Expect = 9e-05
Identities = 23/60 (38%), Positives = 29/60 (48%)
Frame = +2
Query: 119 SPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNPP 178
+P+HPPP P P A P PPP PV P P+PV PS P+ ++ PP
Sbjct: 365 TPYHPPPSPSPPPSPVYAYPSPPP-------PVYTSPPPSPVYAYPSPPPPVYSSPPPPP 523
Score = 40.0 bits (92), Expect = 0.001
Identities = 26/64 (40%), Positives = 27/64 (41%), Gaps = 2/64 (3%)
Frame = +2
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAG-YPVAQVPMPAPV-SIAPSSYVPIQNTKD 175
P P H PPPP PPPPP+ P YP P P PV S P Y P
Sbjct: 128 PPPAHSPPPP-------PXSPPPPPHSPTPPVYPYLSPPPPPPVHSPPPPVYSP--PPPS 280
Query: 176 NPPC 179
PPC
Sbjct: 281 PPPC 292
Score = 39.3 bits (90), Expect = 0.002
Identities = 28/75 (37%), Positives = 32/75 (42%), Gaps = 14/75 (18%)
Frame = +2
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPP--------YDPYAGYP---VAQVPMPAPVSI---A 163
P P H P PPV + Y++PPPPPP Y P P V P P P +
Sbjct: 170 PPPPHSPTPPV---YPYLSPPPPPPVHSPPPPVYSPPPPSPPPCVEPPPPPPPPCVEPPP 340
Query: 164 PSSYVPIQNTKDNPP 178
PSS P Q PP
Sbjct: 341 PSSPAPHQTPYHPPP 385
Score = 37.4 bits (85), Expect = 0.007
Identities = 23/54 (42%), Positives = 26/54 (47%), Gaps = 2/54 (3%)
Frame = +2
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAG--YPVAQVPMPAPVSIAPSSYVP 169
P P PPPPP P +PPPP PY Y+ P A P P P S P + P
Sbjct: 38 PPPNSPPPPPPPAP--VFSPPPPVPY-YYSSPPPPPAHSPPPPPXSPPPPPHSP 190
Score = 35.4 bits (80), Expect = 0.025
Identities = 25/74 (33%), Positives = 33/74 (43%), Gaps = 10/74 (13%)
Frame = +2
Query: 118 PSPFHPPPP-----PVWG-----PHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSY 167
P P + PPP PV+ P+ Y +PPPPP + P P P P P S P Y
Sbjct: 35 PPPPNSPPPPPPPAPVFSPPPPVPYYYSSPPPPPAHSP---PPPPXSPPPPPHSPTPPVY 205
Query: 168 VPIQNTKDNPPCNT 181
P + PP ++
Sbjct: 206 -PYLSPPPPPPVHS 244
Score = 34.7 bits (78), Expect = 0.043
Identities = 22/57 (38%), Positives = 25/57 (43%), Gaps = 14/57 (24%)
Frame = +2
Query: 115 YVTPSP-----FHPPPPPVWG-----PHGYMAPPPPPPYD----PYAGYPVAQVPMP 157
Y PSP PPP PV+ P Y +PPPPP Y+ P G A P P
Sbjct: 413 YAYPSPPPPVYTSPPPSPVYAYPSPPPPVYSSPPPPPVYEGPIPPVFGISYASPPPP 583
Score = 32.7 bits (73), Expect = 0.16
Identities = 23/68 (33%), Positives = 27/68 (38%), Gaps = 8/68 (11%)
Frame = +2
Query: 119 SPFHPPPPPVWGPHGYMAPP---PPPPYDPYAGY-----PVAQVPMPAPVSIAPSSYVPI 170
SP P PPP P PP PPPP P P + P P+PV PS P+
Sbjct: 263 SPPPPSPPPCVEPPPPPPPPCVEPPPPSSPAPHQTPYHPPPSPSPPPSPVYAYPSPPPPV 442
Query: 171 QNTKDNPP 178
+ P
Sbjct: 443 YTSPPPSP 466
Score = 32.0 bits (71), Expect = 0.28
Identities = 19/41 (46%), Positives = 21/41 (50%), Gaps = 12/41 (29%)
Frame = +2
Query: 115 YVTPSP-----FHPPPPPVW-GPH------GYMAPPPPPPY 143
Y PSP PPPPPV+ GP Y +PPPPP Y
Sbjct: 470 YAYPSPPPPVYSSPPPPPVYEGPIPPVFGISYASPPPPPFY 592
Score = 30.8 bits (68), Expect = 0.62
Identities = 18/48 (37%), Positives = 22/48 (45%)
Frame = +2
Query: 131 PHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNPP 178
P+ +PPPPPP P P PAPV +P VP + PP
Sbjct: 11 PYCVRSPPPPPPNSP------PPPPPPAPV-FSPPPPVPYYYSSPPPP 133
>TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG29221.1
[imported] - Arabidopsis thaliana, partial (41%)
Length = 991
Score = 43.5 bits (101), Expect = 9e-05
Identities = 30/76 (39%), Positives = 33/76 (42%), Gaps = 12/76 (15%)
Frame = +1
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPP------PPYDPYAGYPVAQVPMPAPVSI------ 162
YV PSP P PPP P+ Y +PPPP PP PY YP P P P I
Sbjct: 112 YVYPSPPPPSPPP---PYIYKSPPPPPYVYKSPPPPPYV-YPSPPPPSPPPPYIYKSPPP 279
Query: 163 APSSYVPIQNTKDNPP 178
P Y P +PP
Sbjct: 280 PPYVYTPPPYVYKSPP 327
Score = 39.7 bits (91), Expect = 0.001
Identities = 22/53 (41%), Positives = 25/53 (46%)
Frame = +1
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSY 167
YV PSP P PPP P+ Y +PPPPP Y P P+P P Y
Sbjct: 214 YVYPSPPPPSPPP---PYIYKSPPPPPYVYTPPPYVYKSPPPPSPSPPPPYVY 363
Score = 38.5 bits (88), Expect = 0.003
Identities = 28/75 (37%), Positives = 34/75 (45%), Gaps = 11/75 (14%)
Frame = +1
Query: 115 YVTPSPFHP-PPPPV-------WGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPV---SIA 163
YV PSP P PPPP+ P+ Y +PPPP P PY + + P P P S
Sbjct: 37 YVYPSPPPPSPPPPIRIQITTRAPPYVYPSPPPPSPPPPY----IYKSPPPPPYVYKSPP 204
Query: 164 PSSYVPIQNTKDNPP 178
P YV +PP
Sbjct: 205 PPPYVYPSPPPPSPP 249
Score = 35.0 bits (79), Expect = 0.033
Identities = 21/64 (32%), Positives = 26/64 (39%)
Frame = +1
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
Y +P P P PPP P+ Y +PPPPP P P P S P
Sbjct: 313 YKSPPPPSPSPPP---PYVYKSPPPPPYVYKSPPPPPYVYESPPPPQYVYKSPPPPPYVY 483
Query: 175 DNPP 178
++PP
Sbjct: 484 ESPP 495
Score = 34.7 bits (78), Expect = 0.043
Identities = 19/52 (36%), Positives = 22/52 (41%), Gaps = 1/52 (1%)
Frame = +1
Query: 118 PSPF-HPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYV 168
P P+ +P PPP P Y+ PPPP Y P P P P YV
Sbjct: 205 PPPYVYPSPPPPSPPPPYIYKSPPPPPYVYTPPPYVYKSPPPPSPSPPPPYV 360
Score = 33.9 bits (76), Expect = 0.073
Identities = 20/58 (34%), Positives = 26/58 (44%), Gaps = 4/58 (6%)
Frame = +1
Query: 115 YVTPSP----FHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYV 168
Y +P P + PPPP P+ Y +PPPP P P P P+P P Y+
Sbjct: 421 YESPPPPQYVYKSPPPP---PYVYESPPPPSPSPP---PPYYYKPIPPPYVYESPPYI 576
Score = 33.9 bits (76), Expect = 0.073
Identities = 23/73 (31%), Positives = 29/73 (39%), Gaps = 12/73 (16%)
Frame = +1
Query: 118 PSPF---HPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAP---------VSIAPS 165
P P+ PPPP P Y P PPPY + + + P PAP P+
Sbjct: 466 PPPYVYESPPPPSPSPPPPYYYKPIPPPYVYESPPYIYKSPPPAPYVYKSPPYVYKSPPT 645
Query: 166 SYVPIQNTKDNPP 178
Y P Q + PP
Sbjct: 646 PYKPYQYSSPPPP 684
Score = 33.5 bits (75), Expect = 0.095
Identities = 27/72 (37%), Positives = 31/72 (42%), Gaps = 8/72 (11%)
Frame = +1
Query: 115 YVTPSPFHPPPPPVWGPHGYM-APPPPPPYDPYAGYPVAQVPMPAPV--SIAPSSYV--- 168
Y+ SP PPPP V+ P Y+ PPPP P Y P P V S P YV
Sbjct: 256 YIYKSP--PPPPYVYTPPPYVYKSPPPPSPSPPPPYVYKSPPPPPYVYKSPPPPPYVYES 429
Query: 169 --PIQNTKDNPP 178
P Q +PP
Sbjct: 430 PPPPQYVYKSPP 465
Score = 28.1 bits (61), Expect = 4.0
Identities = 10/26 (38%), Positives = 15/26 (57%)
Frame = +1
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPP 140
Y +P + PP + P+ Y +PPPP
Sbjct: 607 YKSPPYVYKSPPTPYKPYQYSSPPPP 684
Score = 27.7 bits (60), Expect = 5.2
Identities = 19/58 (32%), Positives = 21/58 (35%), Gaps = 10/58 (17%)
Frame = +1
Query: 131 PHGYMAPPPPPPYDPYA----------GYPVAQVPMPAPVSIAPSSYVPIQNTKDNPP 178
P+ Y +PPPP P P YP P P P I S P K PP
Sbjct: 34 PYVYPSPPPPSPPPPIRIQITTRAPPYVYPSPPPPSPPPPYIYKSPPPPPYVYKSPPP 207
>BE998536 similar to GP|2598593|emb| MtN12 {Medicago truncatula}, partial
(64%)
Length = 448
Score = 42.4 bits (98), Expect = 2e-04
Identities = 23/52 (44%), Positives = 30/52 (57%), Gaps = 10/52 (19%)
Frame = +3
Query: 118 PSP-FHPPPPPV--WGPH---GYMAPPPP----PPYDPYAGYPVAQVPMPAP 159
P P +H PPPPV + PH GY +PPPP PP+ P+ GY P+ +P
Sbjct: 105 PKPVYHWPPPPVHTYVPHPKPGYHSPPPPGHTYPPHGPHPGYHSPPAPVDSP 260
Score = 33.5 bits (75), Expect = 0.095
Identities = 21/53 (39%), Positives = 23/53 (42%), Gaps = 7/53 (13%)
Frame = +3
Query: 112 HTGYVTPSP-FHPPPPP-----VWGPH-GYMAPPPPPPYDPYAGYPVAQVPMP 157
HT P P +H PPPP GPH GY +PP P P Y P P
Sbjct: 141 HTYVPHPKPGYHSPPPPGHTYPPHGPHPGYHSPPAPVDSPPTPDYYYKSPPPP 299
>AJ498860 similar to GP|20197423|gb|A unknown protein {Arabidopsis thaliana},
partial (80%)
Length = 596
Score = 42.0 bits (97), Expect = 3e-04
Identities = 25/62 (40%), Positives = 27/62 (43%), Gaps = 5/62 (8%)
Frame = +1
Query: 122 HPP---PPPVWGPHGYMAPPPPPPYDPYAGYPVAQ--VPMPAPVSIAPSSYVPIQNTKDN 176
HPP PP + P GY PPP P GYP Q P P P P Y P Q +
Sbjct: 100 HPPQGYPPQGYPPQGYPPQGYPPPGYPPPGYPPQQGYPPPPYPAQGYPPQYAPPQYAQPP 279
Query: 177 PP 178
PP
Sbjct: 280 PP 285
Score = 29.3 bits (64), Expect = 1.8
Identities = 14/37 (37%), Positives = 16/37 (42%), Gaps = 6/37 (16%)
Frame = +1
Query: 110 YNHTGYVTPSPFHPPP------PPVWGPHGYMAPPPP 140
Y GY + PPP PP + P Y PPPP
Sbjct: 175 YPPPGYPPQQGYPPPPYPAQGYPPQYAPPQYAQPPPP 285
>TC87475 similar to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (16%)
Length = 396
Score = 42.0 bits (97), Expect = 3e-04
Identities = 22/53 (41%), Positives = 26/53 (48%)
Frame = +3
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSY 167
Y +P P P PPP P+GY +PPPP P P Y P P+P P Y
Sbjct: 78 YKSPPPPSPSPPP---PYGYKSPPPPSP-SPPPPYIYKSPPSPSPPPYHPYLY 224
Score = 37.7 bits (86), Expect = 0.005
Identities = 18/35 (51%), Positives = 21/35 (59%), Gaps = 2/35 (5%)
Frame = +3
Query: 114 GYVTPSPFHPPPPPVWGPHGYMAP--PPPPPYDPY 146
GY +P P P PPP P+ Y +P P PPPY PY
Sbjct: 123 GYKSPPPPSPSPPP---PYIYKSPPSPSPPPYHPY 218
Score = 37.4 bits (85), Expect = 0.007
Identities = 25/74 (33%), Positives = 33/74 (43%), Gaps = 10/74 (13%)
Frame = +3
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPP-----PPPYDPYAGYPVAQVPMP-----APVSIAP 164
Y +P P PPP P+ Y +PPP PPPY + P + P P +P S +P
Sbjct: 30 YKSPPPPSASPPP---PYYYKSPPPPSPSPPPPYGYKSPPPPSPSPPPPYIYKSPPSPSP 200
Query: 165 SSYVPIQNTKDNPP 178
Y P + PP
Sbjct: 201 PPYHPYLYSSPPPP 242
Score = 35.4 bits (80), Expect = 0.025
Identities = 22/65 (33%), Positives = 28/65 (42%), Gaps = 4/65 (6%)
Frame = +3
Query: 118 PSPFHPPPPPVWG---PHGYMAPPPPPPYDPYA-GYPVAQVPMPAPVSIAPSSYVPIQNT 173
P + PPPP P+ Y +PPPP P P GY P P+P P Y+
Sbjct: 21 PYVYKSPPPPSASPPPPYYYKSPPPPSPSPPPPYGYKSPPPPSPSP----PPPYIYKSPP 188
Query: 174 KDNPP 178
+PP
Sbjct: 189 SPSPP 203
Score = 30.8 bits (68), Expect = 0.62
Identities = 15/35 (42%), Positives = 18/35 (50%)
Frame = +3
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGY 149
Y+ SP P PPP H Y+ PPPP +GY
Sbjct: 168 YIYKSPPSPSPPPY---HPYLYSSPPPPIY*ASGY 263
Score = 30.8 bits (68), Expect = 0.62
Identities = 15/41 (36%), Positives = 19/41 (45%), Gaps = 10/41 (24%)
Frame = +3
Query: 110 YNHTGYVTPSPFHPPP----------PPVWGPHGYMAPPPP 140
Y + PSP PPP PP + P+ Y +PPPP
Sbjct: 120 YGYKSPPPPSPSPPPPYIYKSPPSPSPPPYHPYLYSSPPPP 242
>TC78767 similar to PIR|S59117|S59117 small nuclear ribonucleoprotein U1A -
potato, partial (93%)
Length = 1455
Score = 41.6 bits (96), Expect = 3e-04
Identities = 30/95 (31%), Positives = 45/95 (46%), Gaps = 6/95 (6%)
Frame = +3
Query: 177 PPCNTLFIGNLGENINEEEVRGLFSVQPGFKQMKIIRQERHTVCFIEFEDVNSATNVHHN 236
PP N LFI NL ++ LF PGFK+++++ + + + F+E+ D +T
Sbjct: 948 PPNNILFIQNLPNETTPMMLQMLFLQYPGFKEVRMV-EAKPGIAFVEYGDEMQSTMAMQA 1124
Query: 237 LQGAVIPSSGSIGMRIQYSKN------PFGKRKDG 265
LQG I M I Y+K +G RK G
Sbjct: 1125 LQGFKIAPQNP--MLITYAKK*TVISPTYGARKAG 1223
Score = 37.0 bits (84), Expect = 0.009
Identities = 32/109 (29%), Positives = 52/109 (47%), Gaps = 8/109 (7%)
Frame = +3
Query: 167 YVPIQNTKDNPPCN-TLFIGNLGENINEEEVR----GLFSVQPGFKQMKIIRQERHT-VC 220
Y+ N PP N T++I NL E I +E++ +FS ++ + +H
Sbjct: 444 YMAEVNANPEPPQNMTIYINNLNEKIKIDELKKSLHAVFSQFGKILEVLAFKTLKHKGQA 623
Query: 221 FIEFEDVNSATNVHHNLQGAVIPSSGSIGMRIQY--SKNPFGKRKDGTY 267
++ FEDV SA+N +QG MRIQY +K+ + +GT+
Sbjct: 624 WVIFEDVTSASNALRQMQGFPFYDK---PMRIQYARTKSDVIAKAEGTF 761
>TC76445 similar to PIR|T07623|T07623 extensin homolog HRGP2 - soybean
(fragment), partial (76%)
Length = 821
Score = 41.6 bits (96), Expect = 3e-04
Identities = 22/64 (34%), Positives = 28/64 (43%)
Frame = +2
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
Y +P P P PPP + Y PPPP P Y P P+P+S P+ Y
Sbjct: 122 YKSPPPPSPSPPPPY----YYKSPPPPSPSPPPPYHYQSPPPPSPISHPPNYYKSPPPPS 289
Query: 175 DNPP 178
+PP
Sbjct: 290 PSPP 301
Score = 40.8 bits (94), Expect = 6e-04
Identities = 25/65 (38%), Positives = 29/65 (44%), Gaps = 1/65 (1%)
Frame = +2
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYV-PIQNT 173
Y +P P P PPP P+ Y +PPPP P Y P P+P P YV P
Sbjct: 170 YKSPPPPSPSPPP---PYHYQSPPPPSPISHPPNY-YKSPPPPSPSPPPPYHYVSPPPPV 337
Query: 174 KDNPP 178
K PP
Sbjct: 338 KSPPP 352
Score = 38.1 bits (87), Expect = 0.004
Identities = 24/66 (36%), Positives = 28/66 (42%), Gaps = 11/66 (16%)
Frame = +2
Query: 118 PSPFH----PPPPPVWGPHGYMAPPPPPPYDPYAGY-------PVAQVPMPAPVSIAPSS 166
P P+H PPP P+ P Y PPPP P Y PV P PA + +P
Sbjct: 203 PPPYHYQSPPPPSPISHPPNYYKSPPPPSPSPPPPYHYVSPPPPVKSPPPPAYIYASPPP 382
Query: 167 YVPIQN 172
PI N
Sbjct: 383 --PIYN 394
Score = 37.4 bits (85), Expect = 0.007
Identities = 18/43 (41%), Positives = 22/43 (50%)
Frame = +2
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMP 157
Y +P P P PPP P+ Y++PPPP P Y A P P
Sbjct: 266 YKSPPPPSPSPPP---PYHYVSPPPPVKSPPPPAYIYASPPPP 385
Score = 36.6 bits (83), Expect = 0.011
Identities = 22/64 (34%), Positives = 27/64 (41%)
Frame = +2
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
Y +P P P PP P+ Y +PPPP P P Y P P+P P Y
Sbjct: 26 YKSPPPPSPSPP---SPYYYKSPPPPSP-SPPPPYYYKSPPPPSPSPPPPYYYKSPPPPS 193
Query: 175 DNPP 178
+PP
Sbjct: 194 PSPP 205
Score = 36.2 bits (82), Expect = 0.015
Identities = 20/54 (37%), Positives = 23/54 (42%), Gaps = 4/54 (7%)
Frame = +2
Query: 118 PSPFH----PPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSY 167
PSP++ PPP P P Y PPPP P Y P P+P P Y
Sbjct: 59 PSPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYHY 220
Score = 32.7 bits (73), Expect = 0.16
Identities = 19/55 (34%), Positives = 24/55 (43%)
Frame = +2
Query: 124 PPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNPP 178
PPPP + Y +PPPP P P + Y P P+P P Y +PP
Sbjct: 8 PPPPYY----YKSPPPPSP-SPPSPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPP 157
Score = 31.2 bits (69), Expect = 0.47
Identities = 17/40 (42%), Positives = 19/40 (47%), Gaps = 3/40 (7%)
Frame = +2
Query: 123 PPPPPVWGPHGYMAPPPP---PPYDPYAGYPVAQVPMPAP 159
P PPP P+ Y +PPPP PP Y P P P P
Sbjct: 2 PSPPP---PYYYKSPPPPSPSPPSPYYYKSPPPPSPSPPP 112
>BQ135691 similar to PIR|E96636|E966 hypothetical protein T7P1.21 [imported]
- Arabidopsis thaliana, partial (4%)
Length = 1215
Score = 41.6 bits (96), Expect = 3e-04
Identities = 23/74 (31%), Positives = 30/74 (40%), Gaps = 10/74 (13%)
Frame = -2
Query: 123 PPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPA-PVSIAPSSYV---------PIQN 172
PPPP PH PPP YDP+ + +P P P+S + Y P+
Sbjct: 878 PPPPTTISPHTITRTPPPTAYDPFTARAIPALPSPH*PISSTLTPYTRAP*RTYTHPVPL 699
Query: 173 TKDNPPCNTLFIGN 186
PP T +I N
Sbjct: 698 PPSAPPSPTRYIEN 657
Score = 29.3 bits (64), Expect = 1.8
Identities = 12/24 (50%), Positives = 13/24 (54%)
Frame = +1
Query: 123 PPPPPVWGPHGYMAPPPPPPYDPY 146
PPPPP PPPPPP P+
Sbjct: 1 PPPPP---------PPPPPPPSPF 45
Score = 26.9 bits (58), Expect = 8.9
Identities = 11/27 (40%), Positives = 13/27 (47%)
Frame = +3
Query: 123 PPPPPVWGPHGYMAPPPPPPYDPYAGY 149
PPPPP PPPPPP + +
Sbjct: 3 PPPPP---------PPPPPPLPLFCSF 56
>TC93037 weakly similar to GP|3004934|gb|AAC09237.1| annexin XIV {Neurospora
crassa}, partial (33%)
Length = 778
Score = 41.2 bits (95), Expect = 5e-04
Identities = 24/70 (34%), Positives = 28/70 (39%), Gaps = 2/70 (2%)
Frame = +3
Query: 108 GDYNHTGYVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQV--PMPAPVSIAPS 165
G Y + PSP+ P P GY APP PPP Y P Q P PA + P
Sbjct: 303 GQYGAPPPIPPSPYGHSP----APGGYGAPPGPPPNQSYGAPPPQQYGPPTPASIGYGPP 470
Query: 166 SYVPIQNTKD 175
+ T D
Sbjct: 471 QIIQWDGTAD 500
Score = 37.0 bits (84), Expect = 0.009
Identities = 25/76 (32%), Positives = 29/76 (37%), Gaps = 5/76 (6%)
Frame = +3
Query: 108 GDYNHTGYVTPSPFHPPP---PPVWGPHGYMAPPPPPPYDPYA--GYPVAQVPMPAPVSI 162
G Y G P P PPP PP G + PPP Y Y G P Q P P+
Sbjct: 156 GQYPPQGGYYPPPGGPPPGQYPPQGGHYPPPQGPPPGQYGQYPPQGPPPGQYGAPPPIPP 335
Query: 163 APSSYVPIQNTKDNPP 178
+P + P PP
Sbjct: 336 SPYGHSPAPGGYGAPP 383
Score = 33.9 bits (76), Expect = 0.073
Identities = 27/75 (36%), Positives = 29/75 (38%), Gaps = 10/75 (13%)
Frame = +3
Query: 114 GYVTPSPF-----HPPPPPVWGPHGYMAPPP--PPP--YDPYAG-YPVAQVPMPAPVSIA 163
GY P P +PPP + P G PPP PPP Y P G YP Q P P
Sbjct: 102 GYGQPPPQGGYGGYPPPGGQYPPQGGYYPPPGGPPPGQYPPQGGHYPPPQGPPPGQYGQY 281
Query: 164 PSSYVPIQNTKDNPP 178
P P PP
Sbjct: 282 PPQGPPPGQYGAPPP 326
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 41.2 bits (95), Expect = 5e-04
Identities = 20/47 (42%), Positives = 22/47 (46%), Gaps = 5/47 (10%)
Frame = -3
Query: 118 PSPFHPPPPPVWGP-----HGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
P P+ PPPPP+ P PPPPPPY P P P P P
Sbjct: 788 PPPYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP*PRRSPPYPPP 648
Score = 34.3 bits (77), Expect = 0.056
Identities = 18/44 (40%), Positives = 20/44 (44%), Gaps = 1/44 (2%)
Frame = -3
Query: 117 TPSPFHPPPPPVWGPHGYMA-PPPPPPYDPYAGYPVAQVPMPAP 159
+P P PPPP P + PPPPP PY P P P P
Sbjct: 812 SPPPLLYPPPPYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP 681
Score = 32.3 bits (72), Expect = 0.21
Identities = 18/41 (43%), Positives = 20/41 (47%)
Frame = -3
Query: 119 SPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
SP+ PPPPP + P PPP P P YP P P P
Sbjct: 728 SPYPPPPPPPYPP-----PPP*PRRSP--PYPPPPPP*PPP 627
Score = 31.2 bits (69), Expect = 0.47
Identities = 13/23 (56%), Positives = 14/23 (60%)
Frame = -3
Query: 119 SPFHPPPPPVWGPHGYMAPPPPP 141
SP +PPPPP P PPPPP
Sbjct: 668 SPPYPPPPPP*PPPPRPPPPPPP 600
Score = 30.0 bits (66), Expect = 1.1
Identities = 13/23 (56%), Positives = 14/23 (60%)
Frame = -3
Query: 120 PFHPPPPPVWGPHGYMAPPPPPP 142
P+ PPPPP P PPPPPP
Sbjct: 662 PYPPPPPP--*PPPPRPPPPPPP 600
Score = 29.6 bits (65), Expect = 1.4
Identities = 17/41 (41%), Positives = 18/41 (43%), Gaps = 1/41 (2%)
Frame = -3
Query: 118 PSPFHP-PPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMP 157
P P P PPPP PPPPPP P P P+P
Sbjct: 716 PPPPPPYPPPPP*PRRSPPYPPPPPP*PPPPRPPPPPPPLP 594
Score = 29.3 bits (64), Expect = 1.8
Identities = 18/56 (32%), Positives = 22/56 (39%)
Frame = -3
Query: 123 PPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNPP 178
P PPP+ P PPPPP P + P P P P P ++ PP
Sbjct: 815 PSPPPLLYPPPPYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP*PRRSPPYPPP 648
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 41.2 bits (95), Expect = 5e-04
Identities = 20/47 (42%), Positives = 22/47 (46%), Gaps = 5/47 (10%)
Frame = -2
Query: 118 PSPFHPPPPPVWGP-----HGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
P P+ PPPPP+ P PPPPPPY P P P P P
Sbjct: 534 PPPYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP*PRRSPPYPPP 394
Score = 34.3 bits (77), Expect = 0.056
Identities = 18/44 (40%), Positives = 20/44 (44%), Gaps = 1/44 (2%)
Frame = -2
Query: 117 TPSPFHPPPPPVWGPHGYMA-PPPPPPYDPYAGYPVAQVPMPAP 159
+P P PPPP P + PPPPP PY P P P P
Sbjct: 558 SPPPLLYPPPPYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP 427
Score = 32.3 bits (72), Expect = 0.21
Identities = 18/41 (43%), Positives = 20/41 (47%)
Frame = -2
Query: 119 SPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
SP+ PPPPP + P PPP P P YP P P P
Sbjct: 474 SPYPPPPPPPYPP-----PPP*PRRSP--PYPPPPPP*PPP 373
Score = 31.2 bits (69), Expect = 0.47
Identities = 13/23 (56%), Positives = 14/23 (60%)
Frame = -2
Query: 119 SPFHPPPPPVWGPHGYMAPPPPP 141
SP +PPPPP P PPPPP
Sbjct: 414 SPPYPPPPPP*PPPPRPPPPPPP 346
Score = 30.0 bits (66), Expect = 1.1
Identities = 13/23 (56%), Positives = 14/23 (60%)
Frame = -2
Query: 120 PFHPPPPPVWGPHGYMAPPPPPP 142
P+ PPPPP P PPPPPP
Sbjct: 408 PYPPPPPP--*PPPPRPPPPPPP 346
Score = 29.6 bits (65), Expect = 1.4
Identities = 17/41 (41%), Positives = 18/41 (43%), Gaps = 1/41 (2%)
Frame = -2
Query: 118 PSPFHP-PPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMP 157
P P P PPPP PPPPPP P P P+P
Sbjct: 462 PPPPPPYPPPPP*PRRSPPYPPPPPP*PPPPRPPPPPPPLP 340
Score = 29.3 bits (64), Expect = 1.8
Identities = 18/56 (32%), Positives = 22/56 (39%)
Frame = -2
Query: 123 PPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNPP 178
P PPP+ P PPPPP P + P P P P P ++ PP
Sbjct: 561 PSPPPLLYPPPPYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP*PRRSPPYPPP 394
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 41.2 bits (95), Expect = 5e-04
Identities = 20/47 (42%), Positives = 22/47 (46%), Gaps = 5/47 (10%)
Frame = -2
Query: 118 PSPFHPPPPPVWGP-----HGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
P P+ PPPPP+ P PPPPPPY P P P P P
Sbjct: 692 PPPYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP*PRRSPPYPPP 552
Score = 34.3 bits (77), Expect = 0.056
Identities = 18/44 (40%), Positives = 20/44 (44%), Gaps = 1/44 (2%)
Frame = -2
Query: 117 TPSPFHPPPPPVWGPHGYMA-PPPPPPYDPYAGYPVAQVPMPAP 159
+P P PPPP P + PPPPP PY P P P P
Sbjct: 716 SPPPLLYPPPPYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP 585
Score = 32.3 bits (72), Expect = 0.21
Identities = 18/41 (43%), Positives = 20/41 (47%)
Frame = -2
Query: 119 SPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
SP+ PPPPP + P PPP P P YP P P P
Sbjct: 632 SPYPPPPPPPYPP-----PPP*PRRSP--PYPPPPPP*PPP 531
Score = 31.2 bits (69), Expect = 0.47
Identities = 13/23 (56%), Positives = 14/23 (60%)
Frame = -2
Query: 119 SPFHPPPPPVWGPHGYMAPPPPP 141
SP +PPPPP P PPPPP
Sbjct: 572 SPPYPPPPPP*PPPPRPPPPPPP 504
Score = 30.0 bits (66), Expect = 1.1
Identities = 13/23 (56%), Positives = 14/23 (60%)
Frame = -2
Query: 120 PFHPPPPPVWGPHGYMAPPPPPP 142
P+ PPPPP P PPPPPP
Sbjct: 566 PYPPPPPP--*PPPPRPPPPPPP 504
Score = 29.6 bits (65), Expect = 1.4
Identities = 17/41 (41%), Positives = 18/41 (43%), Gaps = 1/41 (2%)
Frame = -2
Query: 118 PSPFHP-PPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMP 157
P P P PPPP PPPPPP P P P+P
Sbjct: 620 PPPPPPYPPPPP*PRRSPPYPPPPPP*PPPPRPPPPPPPLP 498
Score = 29.3 bits (64), Expect = 1.8
Identities = 18/56 (32%), Positives = 22/56 (39%)
Frame = -2
Query: 123 PPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNPP 178
P PPP+ P PPPPP P + P P P P P ++ PP
Sbjct: 719 PSPPPLLYPPPPYPPPPPPLL*PPPPPRRSPYPPPPPPPYPPPPP*PRRSPPYPPP 552
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.138 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,097,073
Number of Sequences: 36976
Number of extensions: 233353
Number of successful extensions: 9118
Number of sequences better than 10.0: 566
Number of HSP's better than 10.0 without gapping: 2901
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5426
length of query: 284
length of database: 9,014,727
effective HSP length: 95
effective length of query: 189
effective length of database: 5,502,007
effective search space: 1039879323
effective search space used: 1039879323
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144503.7


