

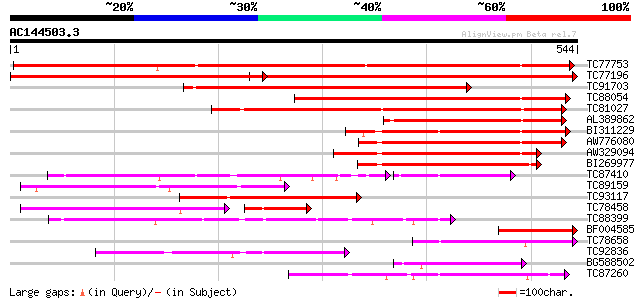
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.3 + phase: 0
(544 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77753 similar to GP|19773582|gb|AAL98709.1 4-coumarate:coenzym... 730 0.0
TC77196 similar to GP|17063848|gb|AAL35216.1 4-coumarate:CoA lig... 582 e-166
TC91703 similar to GP|4038975|gb|AAC97600.1| 4-coumarate:CoA lig... 463 e-131
TC88054 homologue to PIR|PQ0772|PQ0772 4-coumarate--CoA ligase (... 382 e-106
TC81027 similar to PIR|H85064|H85064 4-coumarate--CoA ligase-lik... 276 2e-74
AL389862 similar to GP|23315116|gb Sequence 281 from patent US 6... 209 2e-54
BI311229 similar to GP|20502991|gb Putative AMP-binding protein ... 194 6e-50
AW776080 similar to GP|20161027|db putative 4-coumarate-CoA liga... 194 7e-50
AW329094 similar to GP|20502991|gb Putative AMP-binding protein ... 182 3e-46
BI269977 similar to GP|12039389|gb putative 4-coumarate CoA liga... 160 9e-40
TC87410 similar to GP|16797908|gb|AAL29212.1 putative acyl-CoA s... 89 4e-36
TC89159 weakly similar to PIR|H85064|H85064 4-coumarate--CoA lig... 145 4e-35
TC93117 similar to PIR|H85064|H85064 4-coumarate--CoA ligase-lik... 131 6e-31
TC78458 similar to GP|14532846|gb|AAK64105.1 unknown protein {Ar... 96 3e-29
TC88399 weakly similar to GP|18491217|gb|AAL69511.1 putative AMP... 112 3e-25
BF004585 similar to GP|4038975|gb|A 4-coumarate:CoA ligase isoen... 112 3e-25
TC78658 similar to PIR|A96683|A96683 hypothetical protein F12P19... 98 7e-21
TC92836 weakly similar to GP|20161028|dbj|BAB89961. putative 4-c... 98 7e-21
BG588502 SP|P31552|CAIC Probable crotonobetaine/carnitine-CoA li... 98 7e-21
TC87260 similar to GP|18086379|gb|AAL57649.1 AT3g16910/K14A17_3 ... 92 4e-19
>TC77753 similar to GP|19773582|gb|AAL98709.1 4-coumarate:coenzyme A ligase
{Glycine max}, partial (94%)
Length = 2034
Score = 730 bits (1885), Expect = 0.0
Identities = 362/543 (66%), Positives = 450/543 (82%), Gaps = 4/543 (0%)
Frame = +3
Query: 4 QTMEQELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQ 63
Q +QE IF+SKLPDI IP HLPLHSYCF+NLS++ +RPCLIN T E TY DV LT +
Sbjct: 75 QEQKQEFIFRSKLPDIEIPTHLPLHSYCFQNLSQFNNRPCLINGDTGETLTYSDVHLTVR 254
Query: 64 KVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKAS 123
K+A+GLN LGIQQGDVIM++L N P+F FLGASFRGA++T ANPF+TS+E+AKQA A+
Sbjct: 255 KIAAGLNTLGIQQGDVIMIVLRNSPQFALTFLGASFRGAVITTANPFYTSSELAKQATAT 434
Query: 124 NTKLLVTQACYYDKVKD---LENVKLVFVDSSPEED-NHMHFSELIQADQNEMEEVKVNI 179
+KL++TQ+ Y +K+ D L ++K+V +DSSPEED N + FS L AD+NE+ EVK+N
Sbjct: 435 KSKLIITQSVYLNKINDFAKLIDIKIVCIDSSPEEDENVVDFSVLTNADENELPEVKIN- 611
Query: 180 KPDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHI 239
P+DVVALP+SSGT+GLPKGVMLTH+ LVT+I+Q VDGENP+ Y + EDV+LCVLPMFHI
Sbjct: 612 -PNDVVALPFSSGTSGLPKGVMLTHENLVTTISQLVDGENPHQYTNYEDVLLCVLPMFHI 788
Query: 240 YSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYD 299
Y+LNS+LLCG+R+ A++L++ KF+I L+ KYKVT+A VPPIVLA+ KS E +YD
Sbjct: 789 YALNSILLCGIRSGAAVLIVEKFEITKVLELIEKYKVTVASFVPPIVLALVKSGESMRYD 968
Query: 300 LSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSG 359
LSSIRV+ +G AP+G ELE V+ + P+ LGQGYGMTEAGP L++ L+FAKEP K G
Sbjct: 969 LSSIRVMITGAAPMGMELEQAVKDRLPRTVLGQGYGMTEAGP-LSISLAFAKEPFKTKPG 1145
Query: 360 ACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHT 419
ACGTVVRNAEMKIVD + +SLPRN+ GEICIRG ++MKGYLN+PEAT+ TIDKEGWLHT
Sbjct: 1146ACGTVVRNAEMKIVDTETGASLPRNKAGEICIRGTKVMKGYLNDPEATKRTIDKEGWLHT 1325
Query: 420 GDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEV 479
GDIG IDDDDELFIVDRLKELIKYKG+QVAPAELEA++++HP ISD AVVP+ DEAAGEV
Sbjct: 1326GDIGLIDDDDELFIVDRLKELIKYKGYQVAPAELEALLIAHPNISDAAVVPLKDEAAGEV 1505
Query: 480 PVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLA 539
PVAFVVRSNGS +ED+IK+++S+QVVFYKRINRV+F + IPK+ SGKILRK+L A+L
Sbjct: 1506PVAFVVRSNGS-KISEDEIKQYISQQVVFYKRINRVYFTETIPKAASGKILRKELTARLN 1682
Query: 540 AGV 542
G+
Sbjct: 1683EGL 1691
>TC77196 similar to GP|17063848|gb|AAL35216.1 4-coumarate:CoA ligase {Amorpha
fruticosa}, partial (93%)
Length = 1924
Score = 582 bits (1500), Expect = e-166
Identities = 297/314 (94%), Positives = 302/314 (95%)
Frame = +3
Query: 231 LCVLPMFHIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIA 290
+C + ++ + S LRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIA
Sbjct: 804 MCSSHVSYLLTQLSFAFVVLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIA 983
Query: 291 KSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFA 350
KSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFA
Sbjct: 984 KSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFA 1163
Query: 351 KEPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRET 410
KEPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRET
Sbjct: 1164 KEPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRET 1343
Query: 411 IDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVP 470
IDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVP
Sbjct: 1344 IDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVP 1523
Query: 471 MLDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKIL 530
MLDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKIL
Sbjct: 1524 MLDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKIL 1703
Query: 531 RKDLRAKLAAGVPN 544
RKDLRAKLAAGVPN
Sbjct: 1704 RKDLRAKLAAGVPN 1745
Score = 497 bits (1279), Expect = e-141
Identities = 247/247 (100%), Positives = 247/247 (100%)
Frame = +1
Query: 1 MTIQTMEQELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQL 60
MTIQTMEQELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQL
Sbjct: 112 MTIQTMEQELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQL 291
Query: 61 TAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQA 120
TAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQA
Sbjct: 292 TAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQA 471
Query: 121 KASNTKLLVTQACYYDKVKDLENVKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIK 180
KASNTKLLVTQACYYDKVKDLENVKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIK
Sbjct: 472 KASNTKLLVTQACYYDKVKDLENVKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIK 651
Query: 181 PDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIY 240
PDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIY
Sbjct: 652 PDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIY 831
Query: 241 SLNSVLL 247
SLNSVLL
Sbjct: 832 SLNSVLL 852
>TC91703 similar to GP|4038975|gb|AAC97600.1| 4-coumarate:CoA ligase
isoenzyme 2 {Glycine max}, partial (48%)
Length = 828
Score = 463 bits (1191), Expect = e-131
Identities = 232/278 (83%), Positives = 254/278 (90%), Gaps = 1/278 (0%)
Frame = +1
Query: 167 ADQNE-MEEVKVNIKPDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYH 225
AD+N+ + + K I+PDDVVALPYSSGTTGLPKGVMLTHKGLV+SIAQQVDGENPNLYY
Sbjct: 1 ADENDHLPDAK--IQPDDVVALPYSSGTTGLPKGVMLTHKGLVSSIAQQVDGENPNLYYR 174
Query: 226 SEDVILCVLPMFHIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPI 285
SEDVILCVLP+FHIYSLNSVLLCGLRAKA+ILLMPKFDIN+F LV K+ VT+APVVPPI
Sbjct: 175 SEDVILCVLPLFHIYSLNSVLLCGLRAKATILLMPKFDINSFLNLVNKHGVTVAPVVPPI 354
Query: 286 VLAIAKSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTM 345
VLAIAKSP+L+KYDLSSIR+LKSGGAPLGKELEDTVR KFP A LGQGY MTEAGPVLTM
Sbjct: 355 VLAIAKSPDLNKYDLSSIRILKSGGAPLGKELEDTVRTKFPNAILGQGYRMTEAGPVLTM 534
Query: 346 CLSFAKEPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPE 405
L+FAKEP++VK+ ACGTVVRNAEMKIVDP SLPRNQ GEICIRGDQIMKGYLN+ E
Sbjct: 535 SLAFAKEPLNVKASACGTVVRNAEMKIVDPDTGKSLPRNQSGEICIRGDQIMKGYLNDLE 714
Query: 406 ATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELIKY 443
AT TIDKEGWL+TGDIG+ID+DDELFIVDRLKELIKY
Sbjct: 715 ATERTIDKEGWLYTGDIGYIDEDDELFIVDRLKELIKY 828
>TC88054 homologue to PIR|PQ0772|PQ0772 4-coumarate--CoA ligase (EC
6.2.1.12) (clone GM4CL1B) - soybean (fragment), partial
(62%)
Length = 1055
Score = 382 bits (980), Expect = e-106
Identities = 183/265 (69%), Positives = 228/265 (85%)
Frame = +1
Query: 274 YKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQG 333
+ VT+A VVPP+VLA+AKSP + ++DLSSIR++ SG APLGKELE+T+ + P+A LGQG
Sbjct: 1 HTVTVAMVVPPLVLALAKSPSVAEFDLSSIRLVLSGAAPLGKELEETLHNRIPQAVLGQG 180
Query: 334 YGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRG 393
YGMTEAGPVL+M L FAK P SG+CGTVVRNAE+K++DP+ SL NQPGEICIRG
Sbjct: 181 YGMTEAGPVLSMSLGFAKNPFPTSSGSCGTVVRNAELKVLDPETGRSLGYNQPGEICIRG 360
Query: 394 DQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAEL 453
QIMKGYLN+ AT+ TID+EGWLHTGD+G+IDD+DE+FIVDR+KELIK+KGFQV PAEL
Sbjct: 361 QQIMKGYLNDENATKTTIDEEGWLHTGDVGYIDDNDEIFIVDRVKELIKFKGFQVPPAEL 540
Query: 454 EAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRIN 513
E +++SHP I+D AVVP D AAGEVPVAFVVRSNG+ D TE+ +K+F++KQVVFYKR++
Sbjct: 541 EGLLVSHPSIADAAVVPQKDVAAGEVPVAFVVRSNGN-DLTEEIVKEFIAKQVVFYKRLH 717
Query: 514 RVFFIDAIPKSPSGKILRKDLRAKL 538
+V+F+ AIPKSP+GKILRKDL+AKL
Sbjct: 718 KVYFVHAIPKSPAGKILRKDLKAKL 792
>TC81027 similar to PIR|H85064|H85064 4-coumarate--CoA ligase-like protein
[imported] - Arabidopsis thaliana, partial (28%)
Length = 1283
Score = 276 bits (705), Expect = 2e-74
Identities = 158/345 (45%), Positives = 222/345 (63%), Gaps = 4/345 (1%)
Frame = +1
Query: 194 TGLPKGVMLTHKGLV-TSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVLLCGLRA 252
TG KGVMLTH+ L+ T++A VD + ++V +C +PM H+ L + LR
Sbjct: 1 TGRSKGVMLTHQNLIATAVAGAVDQDQNG---EGKNVFVCFVPMHHVMGLVGITYTQLRR 171
Query: 253 KASILLMP-KFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELD-KYDLSSIRVLKSGG 310
+++ +FD+ F V K+KVT V PP+++ + K E+ Y+LSS++ L G
Sbjct: 172 GNTVVSFGGRFDLEKTFAAVEKFKVTHLYVAPPVMVELIKRREVVIGYELSSLKYLVGGA 351
Query: 311 APLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRNAEM 370
APLGK+L P + QGYGMTEA ++++ I V SG+ GT++ + E
Sbjct: 352 APLGKDLMQECTKILPHVHVIQGYGMTEACGLVSIENPKEGSLISV-SGSTGTLIPSVES 528
Query: 371 KIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDE 430
+I++ LP NQ GEI +RG IM+GY NNPEAT+ I+ +GW+ TGD+G+ D+ +
Sbjct: 529 RIINLATLKPLPPNQLGEIWLRGPTIMQGYFNNPEATKLAINDQGWMITGDLGYFDEKGQ 708
Query: 431 LFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVR-SNG 489
LF+VDR+KELIK G+QVAPAELE +++SHP+ISD V+P D GEVPVAFVVR N
Sbjct: 709 LFVVDRIKELIKCSGYQVAPAELEDLLVSHPEISDAGVIPSPDAKVGEVPVAFVVRLPNS 888
Query: 490 SIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDL 534
SI T++DI+KFV+KQV YKR+ RV F++ IPK +GKILRKDL
Sbjct: 889 SI--TKEDIQKFVAKQVAPYKRLRRVTFLEKIPKLATGKILRKDL 1017
>AL389862 similar to GP|23315116|gb Sequence 281 from patent US 6410718,
partial (90%)
Length = 535
Score = 209 bits (533), Expect = 2e-54
Identities = 107/176 (60%), Positives = 135/176 (75%)
Frame = +1
Query: 359 GACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLH 418
GA G+ V E +IV Q SLP NQ GEI +RG +M+GY NNPEAT++TI+ +GW
Sbjct: 10 GAAGSSV---ESQIVSLQTSKSLPPNQLGEIWLRGPVMMQGYFNNPEATKQTINDQGWTL 180
Query: 419 TGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGE 478
TGD+G+ D+ +LF+VDR+KELIK G+QVAPAELE +++SHP+ISD V+P D AGE
Sbjct: 181 TGDLGYFDEKGQLFVVDRIKELIKCNGYQVAPAELEDLLISHPEISDAGVIPSPDAKAGE 360
Query: 479 VPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDL 534
VPVAFVVRS S+ TE+DIKKFV+K+V YKR+ RV FI+ IPKSP+GKILRKDL
Sbjct: 361 VPVAFVVRSLNSL-ITEEDIKKFVAKEVAPYKRLRRVTFIEKIPKSPTGKILRKDL 525
>BI311229 similar to GP|20502991|gb Putative AMP-binding protein {Oryza
sativa (japonica cultivar-group)}, partial (29%)
Length = 693
Score = 194 bits (494), Expect = 6e-50
Identities = 105/218 (48%), Positives = 146/218 (66%), Gaps = 2/218 (0%)
Frame = +3
Query: 323 AKFPKAKLGQGYGMTE--AGPVLTMCLSFAKEPIDVKSGACGTVVRNAEMKIVDPQNDSS 380
AKFP A + QGYG+TE AG + + A + G G +V E KIV+P +
Sbjct: 39 AKFPHASIIQGYGLTESTAGVIRIVGPEEAS-----RGGTTGKLVSGMEAKIVNPNTGEA 203
Query: 381 LPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKEL 440
+ + GE+ +RG IMKGY+ +P AT T+ +GWL TGDI + D++ +++VDRLKEL
Sbjct: 204 MSPGEQGELWVRGPPIMKGYVGDPVATSVTL-VDGWLRTGDICYFDNEGFVYVVDRLKEL 380
Query: 441 IKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTTEDDIKK 500
IKYKG+QVAPAELE ++ SHP+I D AV+P DE AG++P+AFV+R S E +I
Sbjct: 381 IKYKGYQVAPAELEQLLQSHPEIKDAAVIPYPDEDAGQIPLAFVIRQPHS-SMGEAEIIN 557
Query: 501 FVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKL 538
FV+KQV YK++ RV F+++IPK+ +GKILRKDL K+
Sbjct: 558 FVAKQVAPYKKVRRVVFVNSIPKNATGKILRKDLLNKI 671
>AW776080 similar to GP|20161027|db putative 4-coumarate-CoA ligase {Oryza
sativa (japonica cultivar-group)}, partial (28%)
Length = 625
Score = 194 bits (493), Expect = 7e-50
Identities = 103/200 (51%), Positives = 137/200 (68%)
Frame = +2
Query: 335 GMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGD 394
G+TE+G + F + + G+ G + N E KIVDP +L Q GE+ +RG
Sbjct: 2 GLTESGGGAARMIGFDEAK---RHGSVGRLAENMEAKIVDPVTGEALSPGQKGELWLRGP 172
Query: 395 QIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELE 454
IMKGY+ + +AT ET+D EGWL TGD+ + D D LFIVDRLKELIKYK +QV PAELE
Sbjct: 173 TIMKGYVGDDKATVETLDSEGWLKTGDLCYFDSDGYLFIVDRLKELIKYKAYQVPPAELE 352
Query: 455 AIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINR 514
I+ ++P+I+D AVVP DE AG++P+AFVVR GS + T + +V+KQV YK+I R
Sbjct: 353 HILHTNPEIADAAVVPYPDEDAGQIPMAFVVRKPGS-NITAAQVMDYVAKQVTPYKKIRR 529
Query: 515 VFFIDAIPKSPSGKILRKDL 534
V FI++IPKSP+GKIL K+L
Sbjct: 530 VSFINSIPKSPAGKILIKEL 589
>AW329094 similar to GP|20502991|gb Putative AMP-binding protein {Oryza
sativa (japonica cultivar-group)}, partial (34%)
Length = 595
Score = 182 bits (462), Expect = 3e-46
Identities = 97/200 (48%), Positives = 129/200 (64%)
Frame = +2
Query: 311 APLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRNAEM 370
APL KE+ + K+P + QGYG+TE+ + S + K G G V + E
Sbjct: 8 APLSKEVTEGFIEKYPNVSIFQGYGLTESSGIGASTESLEESR---KYGTAGLVSASTEA 178
Query: 371 KIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDE 430
IVD + LP N+ GE+ +RG MKGY +N EATR TI EGWL TGD+ +ID D
Sbjct: 179 MIVDTETAQPLPVNRTGELWLRGPTTMKGYFSNEEATRSTITPEGWLKTGDVCYIDSDGF 358
Query: 431 LFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGS 490
LF+VDRLKELIKYKG+QV PAELEA++L+HP I D AV+P D+ AG+VP+A+VVR+ GS
Sbjct: 359 LFVVDRLKELIKYKGYQVPPAELEALLLTHPAILDAAVIPYPDKEAGQVPMAYVVRNVGS 538
Query: 491 IDTTEDDIKKFVSKQVVFYK 510
+ + + FV++QV YK
Sbjct: 539 -NLSGSQVMDFVAEQVAPYK 595
>BI269977 similar to GP|12039389|gb putative 4-coumarate CoA ligase {Oryza
sativa}, partial (28%)
Length = 520
Score = 160 bits (406), Expect = 9e-40
Identities = 86/177 (48%), Positives = 118/177 (66%)
Frame = +1
Query: 334 YGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRG 393
YGMTE+G V T + K + G + N E K+VD ++ + LP GE+ +RG
Sbjct: 1 YGMTESGAVGTRGFNTDKFH---NYSSLGLLAPNIEAKVVDWKSGTFLPPGLSGELWLRG 171
Query: 394 DQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAEL 453
IMKGYLNN EAT TIDK+GW+HTGDI + D D L++ RLK++IKYKGFQ+APA+L
Sbjct: 172 PSIMKGYLNNEEATMSTIDKDGWIHTGDIVYFDQDGYLYMSGRLKDIIKYKGFQIAPADL 351
Query: 454 EAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYK 510
EA+++SHP+I D AV + AGE+PVAFVV+ GS+ +++ I +V+ QV YK
Sbjct: 352 EALLISHPEIVDAAVTAGKVDVAGEIPVAFVVKKVGSVLSSQHVI-DYVAXQVAPYK 519
>TC87410 similar to GP|16797908|gb|AAL29212.1 putative acyl-CoA synthetase
{Capsicum annuum}, partial (76%)
Length = 1600
Score = 89.4 bits (220), Expect(2) = 4e-36
Identities = 44/117 (37%), Positives = 69/117 (58%)
Frame = +1
Query: 369 EMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDD 428
EM I+D ++ L + GE+CI+G+ + KGY NN EA + GW HTGDIG+ D D
Sbjct: 1093 EMAILD-ESGRVLEADVNGEVCIKGENVTKGYKNNEEANKSAF-LFGWFHTGDIGYFDSD 1266
Query: 429 DELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVV 485
L +V R+KELI G +++P E++A++L H ++ + D+ GE F++
Sbjct: 1267 GYLHLVGRIKELINRGGEKISPIEVDAVLLGHQDVAQAVAFGVPDQKYGEEVCRFLI 1437
Score = 80.9 bits (198), Expect(2) = 4e-36
Identities = 98/341 (28%), Positives = 148/341 (42%), Gaps = 12/341 (3%)
Frame = +3
Query: 37 KYGSRPCLINAPTAEIYTYYDVQLTAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLG 96
K+ SR + A ++ T+ + + A+ L GI+ DV+ + PN E+V FL
Sbjct: 108 KFPSRRAISVAGKFDL-THSQLHELVESAANHLISAGIKPNDVVALTFPNTIEYVIMFLA 284
Query: 97 ASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVTQACYYDKVKDLE---NVKLVFVDSSP 153
A N +TS E S +KLL+T + + N+ L V +
Sbjct: 285 VIRVRATAAPLNAAYTSEEFEFYLSDSESKLLLTPLEGNNPAQTAATKLNIPLGSVCFNK 464
Query: 154 EEDNHMHFSELIQADQNEMEEVKVNIKPDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQ 213
E+ L Q + +N +P DV ++SGTT PKGV L+ LV+S+
Sbjct: 465 TEEETKLSISLNQTESANSVSELIN-EPSDVALFLHTSGTTSRPKGVPLSQHNLVSSVR- 638
Query: 214 QVDGENPNLYYHSE-DVILCVLPMFHIYSLNSVLLCGLRAKASILL--MPKFDINAFFGL 270
++Y SE D + VLP+FH++ L + LL L A ++ L +F + F+
Sbjct: 639 ----NIESVYRLSELDSTVIVLPLFHVHGLIAGLLSSLGAGGAVALPAAGRFSASTFWKD 806
Query: 271 VTKYKVTIAPVVPPIVLAI----AKSPELDKYDLSSIRVLKSGGAP--LGKELEDTVRAK 324
+ +Y T VP I I +PE L IR + AP LG+ LE+ A
Sbjct: 807 MIQYNATWYTAVPTIHQIILDRHQSNPEPVYPKLRFIRSCSASLAPVILGR-LEEAFGAP 983
Query: 325 FPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVV 365
KA Y MTEA + MC + E K+G+ G V
Sbjct: 984 VLKA-----YAMTEATHL--MCSNPLPEDGPHKAGSVGKPV 1085
>TC89159 weakly similar to PIR|H85064|H85064 4-coumarate--CoA ligase-like
protein [imported] - Arabidopsis thaliana, partial (39%)
Length = 1103
Score = 145 bits (366), Expect = 4e-35
Identities = 88/263 (33%), Positives = 150/263 (56%), Gaps = 5/263 (1%)
Frame = +1
Query: 11 IFKSKLPDIYIPKH--LPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASG 68
I++S P + +PK+ L L ++ F ++ ++ LI+A +++ T+ ++ K+A
Sbjct: 109 IYRSLRPSLILPKNPNLSLVTHLFNKVTSSPNKTALIDADSSQTLTFAQLKSLTIKLAHS 288
Query: 69 LNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLL 128
L K+G+ + DV++ L PN + FL + GA+ + NP +T+ E++KQAK SN KL+
Sbjct: 289 LIKIGLTKNDVVLFLAPNSYLYPVFFLAVASIGAVASTVNPQYTTVEVSKQAKDSNPKLI 468
Query: 129 VTQACYYDKVKDLENVKLVFVDSS--PEEDNHMHFSELIQADQNEMEEVKVNIKPDDVVA 186
+T A ++KV L N+ VF++SS D F ++ ++ E K+++K D A
Sbjct: 469 ITVAELWEKVNHL-NIPAVFLNSSGVATPDTVTSFESFVKLGESATEFPKIDVKQTDTAA 645
Query: 187 LPYSSGTTGLPKGVMLTHKGLVTSIAQ-QVDGENPNLYYHSEDVILCVLPMFHIYSLNSV 245
L YSSGTTG+ KGV+LTH+ + S A +D E L DV L VLPMFH++ L +
Sbjct: 646 LLYSSGTTGVSKGVILTHQNFIASAAMISMDDE---LNGEINDVYLVVLPMFHVFGLAVI 816
Query: 246 LLCGLRAKASILLMPKFDINAFF 268
L+ +I+ + +F+ ++ F
Sbjct: 817 TYSQLQRGNAIVSLKRFEFDSGF 885
>TC93117 similar to PIR|H85064|H85064 4-coumarate--CoA ligase-like protein
[imported] - Arabidopsis thaliana, partial (29%)
Length = 611
Score = 131 bits (330), Expect = 6e-31
Identities = 70/174 (40%), Positives = 105/174 (60%)
Frame = +1
Query: 164 LIQADQNEMEEVKVNIKPDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLY 223
LI++ E + ++K D AL YSSGTTG+ KGV+LTHK + A + + +L
Sbjct: 85 LIESAGEVTELPENDVKQTDTAALLYSSGTTGVSKGVILTHKNFIA--ASLMINMDDDLA 258
Query: 224 YHSEDVILCVLPMFHIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVP 283
+V +CVLPMFH++ L + LR ++++ M +F++ F ++ KYKVT VVP
Sbjct: 259 GELNNVFICVLPMFHVFGLAVITYAQLRRGSTVVSMGRFELEGFLKVIEKYKVTNLWVVP 438
Query: 284 PIVLAIAKSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMT 337
P++LA+AK + KYD+SS++ + SG APLGKEL + P + GYGMT
Sbjct: 439 PMILALAKQSVVGKYDVSSLKSIGSGAAPLGKELMEECAXHLPXVAIYXGYGMT 600
>TC78458 similar to GP|14532846|gb|AAK64105.1 unknown protein {Arabidopsis
thaliana}, partial (49%)
Length = 1066
Score = 95.9 bits (237), Expect(2) = 3e-29
Identities = 63/208 (30%), Positives = 99/208 (47%), Gaps = 7/208 (3%)
Frame = +2
Query: 11 IFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASGLN 70
IF SK + +P + L + F + + I+A T +TY + + V S L+
Sbjct: 194 IFYSKRKPLPLPPNHSLDATTFISSRAHHGHIAFIDASTGHQFTYQQLWRSVDSVTSSLS 373
Query: 71 KLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVT 130
+GI++GDVI++L PN F L GAI+T NP TS EIAKQ S L T
Sbjct: 374 NMGIRKGDVILLLSPNSIYFPVVCLSVMSLGAIITTTNPLNTSGEIAKQIADSKPVLAFT 553
Query: 131 QACYYDKVKDLE-NVKLVFVDSSPEEDNHMHFS------ELIQADQNEMEEVKVNIKPDD 183
K+ ++ ++ +D++ +N S + + + E+ V + +D
Sbjct: 554 TQQLLSKITQASPSLPIILMDTNNNNNNSSSSSTTTTTLDEMMKKEPELSRVTERVNQND 733
Query: 184 VVALPYSSGTTGLPKGVMLTHKGLVTSI 211
L YSSGTTG KGV+ +HK L+ +
Sbjct: 734 TATLLYSSGTTGPSKGVVSSHKNLIAMV 817
Score = 51.2 bits (121), Expect(2) = 3e-29
Identities = 24/65 (36%), Positives = 42/65 (63%), Gaps = 1/65 (1%)
Frame = +3
Query: 226 SEDVILCVLPMFHIYSLNSVLLCGLRAKAS-ILLMPKFDINAFFGLVTKYKVTIAPVVPP 284
+ + +C +PMFHIY L +V GL A S I+++ KF+++ + K++ T P+VPP
Sbjct: 855 TRETFICTVPMFHIYGL-AVFAMGLLAMGSTIVVLSKFEMHDMLSSIEKFRATFLPLVPP 1031
Query: 285 IVLAI 289
I++A+
Sbjct: 1032ILVAM 1046
>TC88399 weakly similar to GP|18491217|gb|AAL69511.1 putative AMP-binding
protein {Arabidopsis thaliana}, partial (62%)
Length = 1286
Score = 112 bits (281), Expect = 3e-25
Identities = 108/420 (25%), Positives = 181/420 (42%), Gaps = 30/420 (7%)
Frame = +3
Query: 38 YGSRPCLINAPTAEIYTYYDVQLTAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGA 97
Y + P +I T ++T+ ++AS + LGI++G+V+ V+ PN P
Sbjct: 75 YPNTPSIIYNDT--VFTWSQTHKRCLQLASAITSLGIRRGNVVSVIAPNIPAMYELHFAV 248
Query: 98 SFRGAIMTAANPFFTSAEIAKQAKASNTKLLVTQACYYDKV------------------- 138
F GAI+ N + I+ +KL+ D V
Sbjct: 249 PFTGAILNNINTRLDARIISNILLHCESKLVFVDIAARDLVLQALSLFPSKQHKPLLILI 428
Query: 139 KDLENVKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKPDDVVALPYSSGTTGLPK 198
KD V SS D + +L+ + V N + D ++ L Y+SGTT PK
Sbjct: 429 KDQTFETYENVSSSSTVDFISTYEDLMASGDPNFNWVYPNSEWDPML-LNYTSGTTSSPK 605
Query: 199 GVMLTHKG-LVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVLLCGLRAKASI- 256
GV+ +H+G + ++ ++ P + + L LPMFH + G+ A
Sbjct: 606 GVVHSHRGAFIVTVDTLIEWTVP-----KQPIYLWTLPMFHANGWS--FPWGIAAVGGTN 764
Query: 257 LLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAPLGKE 316
+ + KFD F L+ K+ VT P ++ + SP+ +K ++ +L +G P
Sbjct: 765 ICVRKFDAEIVFSLIRKHHVTHMCGAPVVLNMLTNSPD-NKPLQKTVHILTAGAPPPAAV 941
Query: 317 LEDTVRAKFPKAKLGQGYGMTEAGPVLTMC-------LSFAKEPIDVKSGACGTVVRNAE 369
L T F + GYG+TE G ++ C L A E +KS + +
Sbjct: 942 LHRTEALGF---TVSHGYGLTETGGLVVCCTWKKKWNLLPATERARLKSRQGVRTLGMTK 1112
Query: 370 MKIVDPQNDSSLPRNQP--GEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDD 427
+ +V P + S+ R+ GE+ +RG +M GYL + E T+ K+GW +TGD+G + +
Sbjct: 1113VDVVGPSGE-SVKRDGATLGEVVMRGGCVMLGYLKDSEGTKNCF-KKGWFYTGDVGVMHE 1286
>BF004585 similar to GP|4038975|gb|A 4-coumarate:CoA ligase isoenzyme 2
{Glycine max}, partial (13%)
Length = 304
Score = 112 bits (281), Expect = 3e-25
Identities = 58/75 (77%), Positives = 63/75 (83%)
Frame = +2
Query: 470 PMLDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKI 529
PM DEAAGEVPVAFVVRSNG D T+D+I F+SK VVF IN+VFFIDAIPKSPS KI
Sbjct: 5 PMNDEAAGEVPVAFVVRSNGYTDLTQDEINHFISKHVVFNTTINQVFFIDAIPKSPSCKI 184
Query: 530 LRKDLRAKLAAGVPN 544
LRKDLRAKLAA VP+
Sbjct: 185 LRKDLRAKLAASVPH 229
>TC78658 similar to PIR|A96683|A96683 hypothetical protein F12P19.5
[imported] - Arabidopsis thaliana, partial (29%)
Length = 813
Score = 98.2 bits (243), Expect = 7e-21
Identities = 60/164 (36%), Positives = 90/164 (54%), Gaps = 6/164 (3%)
Frame = +2
Query: 387 GEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGF 446
GEI ++G IM GY + EAT + +GW TGD+G I D L I DR K++I G
Sbjct: 65 GEIMLKGSGIMMGYFKDKEATAKAFG-DGWFRTGDVGVIHKDGYLEIKDRSKDVIISGGE 241
Query: 447 QVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFV-VRSNGSIDT-----TEDDIKK 500
++ E+E ++ SHP++ + AVV M GE P AFV + N + + TED+I
Sbjct: 242 NISSVEVENVLYSHPKVLEAAVVAMPHPKWGESPCAFVTLNKNEEVKSDFCCVTEDEIIT 421
Query: 501 FVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAGVPN 544
+ K + + V F++ +PK+ +GKI + +LRAK V N
Sbjct: 422 YCRKNLPHFMVPKMVVFMEDLPKTLTGKIQKFELRAKAKCFVVN 553
>TC92836 weakly similar to GP|20161028|dbj|BAB89961. putative
4-coumarate-CoA ligase {Oryza sativa (japonica
cultivar-group)}, partial (12%)
Length = 720
Score = 98.2 bits (243), Expect = 7e-21
Identities = 69/250 (27%), Positives = 112/250 (44%), Gaps = 6/250 (2%)
Frame = +1
Query: 83 LLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVTQACYYDKVKDLE 142
L+P+ + G + ANP + +E++ N + + + K+ L
Sbjct: 1 LIPSSIHVPVLYFSLLSLGVTIAPANPLSSPSELSHLVHLINPVIAFSTSSTATKIPKLP 180
Query: 143 NVKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKPDDVVALPYSSGTTGLPKGVML 202
++F D+ S L + E V D A+ +SSGTTG KGV+L
Sbjct: 181 LGTVIF-------DSPSFLSVLNSTIDTDTELRPVETSQSDTAAILFSSGTTGRVKGVLL 339
Query: 203 THKGLVTSIA------QQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVLLCGLRAKASI 256
TH+ + I ++ P+ V LP+FH+ +++ + ++
Sbjct: 340 THRNFIALIGGFCFLRHGIEDHEPH-----PGVRFFRLPLFHVIGF-FMMVRTMAMGETL 501
Query: 257 LLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAPLGKE 316
+LM +FD V KY++T PV PP++ KS + KYD+SSIR L GGAPL K
Sbjct: 502 VLMQRFDFGGMLKAVEKYRITHMPVSPPLITXFTKSELVKKYDVSSIRSLGCGGAPLAKX 681
Query: 317 LEDTVRAKFP 326
++ +AKFP
Sbjct: 682 GAESFKAKFP 711
>BG588502 SP|P31552|CAIC Probable crotonobetaine/carnitine-CoA ligase (EC
6.3.2.-). {Escherichia coli}, partial (50%)
Length = 807
Score = 98.2 bits (243), Expect = 7e-21
Identities = 55/131 (41%), Positives = 76/131 (57%), Gaps = 3/131 (2%)
Frame = -1
Query: 369 EMKIVDPQNDSSLPRNQPGEICIRG---DQIMKGYLNNPEATRETIDKEGWLHTGDIGFI 425
E +I D N LP + GEICI+G I K Y NP+AT + ++ +GWLHTGD G+
Sbjct: 411 EAEIRDDHN-RPLPAGEIGEICIKGIPGKTIFKEYFLNPQATAKVLEADGWLHTGDTGYR 235
Query: 426 DDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVV 485
D++D + VDR +IK G V+ ELE II +HP+I D+ VV + D E AFVV
Sbjct: 234 DEEDFFYFVDRRCNMIKRGGENVSCVELENIIAAHPKIQDIVVVGIKDSIRDEAIKAFVV 55
Query: 486 RSNGSIDTTED 496
+ G + E+
Sbjct: 54 LNEGETLSEEE 22
>TC87260 similar to GP|18086379|gb|AAL57649.1 AT3g16910/K14A17_3
{Arabidopsis thaliana}, partial (48%)
Length = 1102
Score = 92.4 bits (228), Expect = 4e-19
Identities = 86/288 (29%), Positives = 133/288 (45%), Gaps = 18/288 (6%)
Frame = +3
Query: 268 FGLVTKYKVTIAPVVPPIVLAIAKSP-ELDKYDLSSI-RVLKSGGAPLGKELEDTVRAKF 325
+ + KYKVT P ++ +I +P E L + V +G AP L F
Sbjct: 6 YDAIVKYKVTHFCAAPVVLNSIINAPAEETILPLPHVVHVNTAGAAPPPSVLSGMSERGF 185
Query: 326 PKAKLGQGYGMTEA-GPVLTMCLSFAKEPIDVKSGA---CGTVVRNAEMKIVDPQNDSSL 381
++ YG++E GP + + + +S A V+ ++ +D N ++
Sbjct: 186 ---RVTHTYGLSETYGPSVYCAWKPEWDSLPPESRARLHARQGVKYIALEGLDVVNTKTM 356
Query: 382 PRNQP--------GEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFI 433
QP GEI +RG+ +MKGYL NP+A E+ GW H+GD+ D + I
Sbjct: 357 ---QPVPADGKTVGEIVMRGNAVMKGYLKNPKANEESF-ANGWYHSGDLAVKHPDGYIEI 524
Query: 434 VDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDT 493
DR K++I ++ E+E + SHP I + +VV DE GE P AFV G +
Sbjct: 525 KDRSKDIIISGAENISSVEVENALYSHPAILETSVVARPDEKWGESPCAFVTLKPGVDRS 704
Query: 494 TE----DDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAK 537
E +DI KF ++ Y V F +PK+ +GK+ + LRAK
Sbjct: 705 NEQRLVEDILKFCRAKMPTYWVPKSVVF-GPLPKTATGKVQKHLLRAK 845
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,021,436
Number of Sequences: 36976
Number of extensions: 195028
Number of successful extensions: 1058
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 992
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1002
length of query: 544
length of database: 9,014,727
effective HSP length: 101
effective length of query: 443
effective length of database: 5,280,151
effective search space: 2339106893
effective search space used: 2339106893
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144503.3


