

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.2 + phase: 0
(322 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
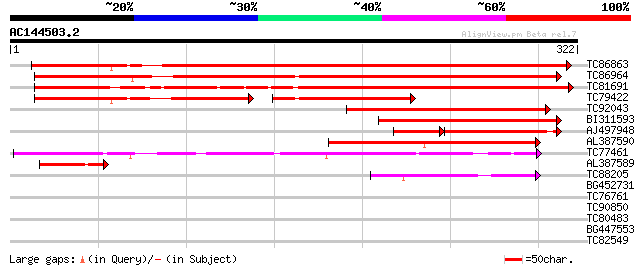
Score E
Sequences producing significant alignments: (bits) Value
TC86863 similar to GP|19387263|gb|AAL87174.1 putative apospory-a... 438 e-123
TC86964 similar to PIR|T47953|T47953 hypothetical protein F2A19.... 348 1e-96
TC81691 weakly similar to GP|6016717|gb|AAF01543.1| unknown prot... 308 2e-84
TC79422 similar to PIR|T47953|T47953 hypothetical protein F2A19.... 118 3e-53
TC92043 similar to GP|21554299|gb|AAM63374.1 apospory-associated... 178 2e-45
BI311593 similar to PIR|T47953|T47 hypothetical protein F2A19.21... 153 7e-38
AJ497948 similar to GP|8777366|dbj| apospory-associated protein ... 99 4e-32
AL387590 weakly similar to GP|19387263|gb| putative apospory-ass... 108 3e-24
TC77461 similar to GP|10177534|dbj|BAB10929. apospory-associated... 96 2e-20
AL387589 weakly similar to GP|19387263|gb| putative apospory-ass... 49 3e-06
TC88205 similar to GP|15028029|gb|AAK76545.1 unknown protein {Ar... 44 1e-04
BG452731 weakly similar to PIR|A96596|A965 hypothetical protein ... 36 0.023
TC76761 similar to GP|19424061|gb|AAL87257.1 unknown protein {Ar... 29 2.8
TC90850 similar to GP|23496018|gb|AAN35680.1 hypothetical protei... 28 4.8
TC80483 similar to GP|22136520|gb|AAM91338.1 putative protein {A... 28 6.2
BG447553 similar to GP|15292785|gb| putative ribosomal-protein S... 28 6.2
TC82549 similar to GP|15292785|gb|AAK92761.1 putative ribosomal-... 28 6.2
>TC86863 similar to GP|19387263|gb|AAL87174.1 putative apospory-associated
protein C {Oryza sativa (japonica cultivar-group)},
partial (88%)
Length = 1640
Score = 438 bits (1126), Expect = e-123
Identities = 211/311 (67%), Positives = 254/311 (80%), Gaps = 4/311 (1%)
Frame = +1
Query: 13 GVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK----PEKEVKKTNQYN 68
G+NGL+K++LRE+RG SAE+YLYGG VTSWKN+ EELLF+SSK P K ++
Sbjct: 244 GINGLDKVLLRESRGASAEIYLYGGHVTSWKNDHAEELLFLSSKAIFKPPKAIRGGIPI- 420
Query: 69 CFDMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSE 128
CF N + + F +N++W ++ +PPPFPTN+ S+AFVDL+LK SE
Sbjct: 421 CFPQFAN-----------HGNLESHGFARNRVWAIEDDPPPFPTNNLSKAFVDLILKPSE 567
Query: 129 DDTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRI 188
+D K W H +E+RLR+ALGP GDLMLTSRIRNTN DGK F+FTFAYHTYL V+DISEVR+
Sbjct: 568 EDMKIWNHSFEYRLRVALGPGGDLMLTSRIRNTNSDGKPFSFTFAYHTYLSVSDISEVRV 747
Query: 189 EGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGL 248
EGLETLDYLD+L+N+ERFTEQGDA+TFE EVDK+YLSTPTKIAIIDHE+KRTF +RKDGL
Sbjct: 748 EGLETLDYLDNLQNKERFTEQGDALTFESEVDKIYLSTPTKIAIIDHEKKRTFVLRKDGL 927
Query: 249 PDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSY 308
PDAVVWNPWDKK+K ++D GDDEYKHMLCV+ A +EKAITLKPGEEWKGR E+SAVPSSY
Sbjct: 928 PDAVVWNPWDKKAKAMADFGDDEYKHMLCVEAANIEKAITLKPGEEWKGRLELSAVPSSY 1107
Query: 309 CSGQLDPRKVL 319
CSGQLDP++VL
Sbjct: 1108CSGQLDPQRVL 1140
>TC86964 similar to PIR|T47953|T47953 hypothetical protein F2A19.210 -
Arabidopsis thaliana, partial (98%)
Length = 1404
Score = 348 bits (894), Expect = 1e-96
Identities = 169/303 (55%), Positives = 222/303 (72%), Gaps = 4/303 (1%)
Frame = +2
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYN---CFD 71
NG+ +++LR +G SA V L G QVTSW+NE+GEELLF SSK + +K + CF
Sbjct: 272 NGVSQVVLRTPQGASARVSLNGAQVTSWRNEQGEELLFTSSKTTPKAQKAARGGIPICFP 451
Query: 72 MNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNS-RAFVDLVLKNSEDD 130
N + L + F +N++W++D NPPP P N +S ++FVDL+LK+SE+D
Sbjct: 452 QFGNCGSMEL-----------HGFARNRMWVIDENPPPLPANDSSGKSFVDLLLKSSEED 598
Query: 131 TKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEG 190
K WPH +EFRLR++L GD+ L SR+RN N GK F+F+FAYHTYL V+DISE+RIEG
Sbjct: 599 MKCWPHSFEFRLRVSLTTNGDITLISRVRNIN--GKPFSFSFAYHTYLLVSDISEIRIEG 772
Query: 191 LETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPD 250
LETLDYLD+L +ERFTEQGDAITFE EVD+VYLS+P IA++DHERK+T+ I+K+GLPD
Sbjct: 773 LETLDYLDNLSQKERFTEQGDAITFESEVDRVYLSSPNIIAVLDHERKQTYIIKKEGLPD 952
Query: 251 AVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYCS 310
VWNPW+KKSK ++D GD+EYK MLCV A +E + L+PGEEW GR ++S VPSS+C
Sbjct: 953 VAVWNPWEKKSKSMTDFGDEEYKQMLCVDAAVIETPVNLRPGEEWTGRLQLSIVPSSFCG 1132
Query: 311 GQL 313
+L
Sbjct: 1133DRL 1141
>TC81691 weakly similar to GP|6016717|gb|AAF01543.1| unknown protein
{Arabidopsis thaliana}, complete
Length = 1465
Score = 308 bits (789), Expect = 2e-84
Identities = 168/306 (54%), Positives = 211/306 (68%)
Frame = +3
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYNCFDMNN 74
+GL +I+L + G SAEV LYGG + SWKN+R EELLF+SSK K Q+
Sbjct: 87 DGLPRIVLTDPNGSSAEVLLYGGHIVSWKNQRKEELLFMSSKA-----KWKQHKAIRGGI 251
Query: 75 NKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDDTKNW 134
+ C D+ Q+ +N++W LD +P P P N + VDL+LK++ D K
Sbjct: 252 SAC--FARFGDV-GSLEQHGMTRNRMWSLDRDPSPLPPIDNHSS-VDLILKSTGVDLKT- 416
Query: 135 PHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEGLETL 194
P +E RLRI+L TG L+L R+RNT D K F+F F+ YL V+DISEVRIEGLETL
Sbjct: 417 PQSFELRLRISLS-TGKLILIPRVRNT--DNKPFSFAFSLCNYLSVSDISEVRIEGLETL 587
Query: 195 DYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPDAVVW 254
DY+D+L NR RFTEQ DAITF+GE D+VYL +P KIAIIDHE+KRTF ++K+ LPDAVVW
Sbjct: 588 DYVDNLMNRSRFTEQADAITFDGETDRVYLHSPNKIAIIDHEKKRTFVLQKNSLPDAVVW 767
Query: 255 NPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYCSGQLD 314
NPW+KK+K + DLG D+YK ML V A ++ I LKP EEWKG QEIS V SSYCSGQLD
Sbjct: 768 NPWNKKAKALPDLGVDDYKMMLRVNSAAIDTPIILKPCEEWKGYQEISTVSSSYCSGQLD 947
Query: 315 PRKVLF 320
PR+VL+
Sbjct: 948 PRRVLY 965
>TC79422 similar to PIR|T47953|T47953 hypothetical protein F2A19.210 -
Arabidopsis thaliana, partial (60%)
Length = 1650
Score = 118 bits (295), Expect(2) = 3e-53
Identities = 63/128 (49%), Positives = 81/128 (63%), Gaps = 4/128 (3%)
Frame = +2
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK----PEKEVKKTNQYNCF 70
NG+E+++LR RG SA V L+GGQV SWK ERGEELLF S+K P K V+ CF
Sbjct: 1067 NGIEQVVLRNVRGASATVSLHGGQVLSWKTERGEELLFTSTKAVYTPPKPVRGGIPI-CF 1243
Query: 71 DMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDD 130
N+ L Q+ F +NK+W++D NPPP PT+SN +A +DL+LK SEDD
Sbjct: 1244 PQFGNRGTLE-----------QHGFARNKIWVIDQNPPPLPTDSNGKACIDLLLKPSEDD 1390
Query: 131 TKNWPHRY 138
K WPHR+
Sbjct: 1391 MKIWPHRW 1414
Score = 108 bits (269), Expect(2) = 3e-53
Identities = 51/81 (62%), Positives = 66/81 (80%)
Frame = +3
Query: 150 GDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEGLETLDYLDSLKNRERFTEQ 209
G L + SRIRN N GK F+F+FA+HTY ++DI+EVR+EGLETLDYLD+L +ERFTEQ
Sbjct: 1413 GRLNMISRIRNVN--GKPFSFSFAFHTYFSISDINEVRVEGLETLDYLDNLHQKERFTEQ 1586
Query: 210 GDAITFEGEVDKVYLSTPTKI 230
GDA+TFE EVD+VYL + + +
Sbjct: 1587 GDALTFESEVDRVYLDSSSMV 1649
>TC92043 similar to GP|21554299|gb|AAM63374.1 apospory-associated protein C
{Arabidopsis thaliana}, partial (37%)
Length = 1163
Score = 178 bits (452), Expect = 2e-45
Identities = 82/116 (70%), Positives = 96/116 (82%)
Frame = +2
Query: 192 ETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPDA 251
ETLDYLD+L +ERFTEQGDA+TFE EVD+VYL + + +A++DHERKRTF IRK+GLPD
Sbjct: 509 ETLDYLDNLHQKERFTEQGDALTFESEVDRVYLDSSSMVAVLDHERKRTFVIRKEGLPDV 688
Query: 252 VVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSS 307
VVWNPW+KKSK I D GD+EYK MLCV A V K ITLKPGEEW GR E+S VPS+
Sbjct: 689 VVWNPWEKKSKSIVDFGDEEYKQMLCVDGAAVGKPITLKPGEEWTGRLELSVVPST 856
>BI311593 similar to PIR|T47953|T47 hypothetical protein F2A19.210 -
Arabidopsis thaliana, partial (33%)
Length = 423
Score = 153 bits (387), Expect = 7e-38
Identities = 68/104 (65%), Positives = 84/104 (80%)
Frame = +1
Query: 210 GDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPDAVVWNPWDKKSKIISDLGD 269
GDA+TFE EVD+VYLS+P IA++DHERKRT+ IRK+GLPD VWNPW+KKSK + D GD
Sbjct: 4 GDAVTFESEVDRVYLSSPNIIAVLDHERKRTYIIRKEGLPDVAVWNPWEKKSKSMVDFGD 183
Query: 270 DEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYCSGQL 313
+EYK MLCV A +EK + LKPGEEW GR ++S V SS+CS +L
Sbjct: 184 EEYKQMLCVDGAVIEKPVNLKPGEEWTGRIQLSVVRSSFCSDRL 315
>AJ497948 similar to GP|8777366|dbj| apospory-associated protein C
{Arabidopsis thaliana}, partial (25%)
Length = 520
Score = 99.0 bits (245), Expect(2) = 4e-32
Identities = 45/66 (68%), Positives = 53/66 (80%)
Frame = +3
Query: 248 LPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSS 307
LPDAVVWNPWDKK+K ++D GDDEYKHMLCV+ A +EKAITLKPGEEWK R + +V
Sbjct: 111 LPDAVVWNPWDKKAKAMADFGDDEYKHMLCVEAANIEKAITLKPGEEWKERSLLCSVS-- 284
Query: 308 YCSGQL 313
C+G L
Sbjct: 285 -CTGSL 299
Score = 56.6 bits (135), Expect(2) = 4e-32
Identities = 25/29 (86%), Positives = 28/29 (96%)
Frame = +1
Query: 219 VDKVYLSTPTKIAIIDHERKRTFEIRKDG 247
VDK+YLSTPTKIAIIDHE+KRTF +RKDG
Sbjct: 25 VDKIYLSTPTKIAIIDHEKKRTFVLRKDG 111
>AL387590 weakly similar to GP|19387263|gb| putative apospory-associated
protein C {Oryza sativa (japonica cultivar-group)},
partial (14%)
Length = 524
Score = 108 bits (270), Expect = 3e-24
Identities = 52/122 (42%), Positives = 76/122 (61%), Gaps = 2/122 (1%)
Frame = +2
Query: 182 DISEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIID--HERKR 239
DIS++ +EGL + Y+D +KN + ++ F E D++Y + PT+I I+ E
Sbjct: 2 DISKISVEGLHNISYIDKVKNFITVVDNDKSVKFTQETDRIYGNVPTEIIKINISEESNE 181
Query: 240 TFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQ 299
F +++ L D+VVWNPW K+K +SD DDEYK M+CV+P V + ITLKP E W+G Q
Sbjct: 182 AFLLQRINLKDSVVWNPWIDKAKGMSDFDDDEYKQMVCVEPGHVTEYITLKPEESWEGGQ 361
Query: 300 EI 301
I
Sbjct: 362 TI 367
>TC77461 similar to GP|10177534|dbj|BAB10929. apospory-associated protein
C-like {Arabidopsis thaliana}, partial (89%)
Length = 1323
Score = 95.5 bits (236), Expect = 2e-20
Identities = 82/307 (26%), Positives = 132/307 (42%), Gaps = 7/307 (2%)
Frame = +3
Query: 3 RETMYVKHCMGVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVK 62
+ T+ VK G L K++L G AE+YL+GG +TSWK G++LLFV +P+
Sbjct: 132 KTTLGVKVTEGEGNLPKLVLTSPAGSEAEIYLFGGCITSWKVPNGKDLLFV--RPDAVFN 305
Query: 63 KTNQY-----NCFDMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSR 117
K +CF Q Q+ F +N W + + N
Sbjct: 306 KKKPISGGIPHCFP------------QFGPGAIQQHGFARNLDWTVGDS-----ENVEGN 434
Query: 118 AFVDLVLKNSEDDTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTY 177
V L LK+ W + ++ L L++ ++ N D K+F+F A HTY
Sbjct: 435 PVVTLELKDDTYSRAMWDFSFHALYKVTLNAKS---LSTELKVKNTDNKAFSFNTALHTY 605
Query: 178 L--YVTDISEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDH 235
V+ S ++G +TL+ N +E+ D +TF G VD VYL+ ++ +D+
Sbjct: 606 FRASVSGASVKGLKGCKTLNKHPDPNNPIEGSEERDVVTFPGFVDCVYLNASDELQ-LDN 782
Query: 236 ERKRTFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEW 295
I+ DAV+WNP + YK +CV+ A + + L+P + W
Sbjct: 783 GLGDIISIKNTNWSDAVLWNPHLQMEAC--------YKDFVCVENAKIGN-VQLEPKQTW 935
Query: 296 KGRQEIS 302
Q ++
Sbjct: 936 TAVQHLT 956
>AL387589 weakly similar to GP|19387263|gb| putative apospory-associated
protein C {Oryza sativa (japonica cultivar-group)},
partial (7%)
Length = 472
Score = 48.9 bits (115), Expect = 3e-06
Identities = 22/39 (56%), Positives = 32/39 (81%)
Frame = +3
Query: 18 EKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK 56
+K+IL++++G SAE+Y +G VTSWK RG+ELLF+S K
Sbjct: 6 DKVILKDSKGSSAEIYYFGATVTSWK-YRGKELLFLSKK 119
>TC88205 similar to GP|15028029|gb|AAK76545.1 unknown protein {Arabidopsis
thaliana}, partial (45%)
Length = 1371
Score = 43.5 bits (101), Expect = 1e-04
Identities = 31/109 (28%), Positives = 51/109 (46%), Gaps = 13/109 (11%)
Frame = +1
Query: 206 FTEQGDAITF-EGEVDKV------------YLSTPTKIAIIDHERKRTFEIRKDGLPDAV 252
+T+QG IT E ++ +V Y + P+K IID R+ + + + G D
Sbjct: 859 WTQQGVPITLLENKMSRVFAAPPKERTKAFYNTPPSKYEIIDQGREIFYRVIRMGFEDIY 1038
Query: 253 VWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEI 301
V +P K D + +C PA + + +T+ PGEE++G Q I
Sbjct: 1039VSSPGSMSDKYGRD-------YFVCTGPASILEPVTVNPGEEFRGAQVI 1164
>BG452731 weakly similar to PIR|A96596|A965 hypothetical protein T18I3.2
[imported] - Arabidopsis thaliana, partial (14%)
Length = 656
Score = 35.8 bits (81), Expect = 0.023
Identities = 20/79 (25%), Positives = 38/79 (47%)
Frame = +3
Query: 219 VDKVYLSTPTKIAIIDHERKRTFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCV 278
+ VY + P +ID R+ + + ++G + +++P + S +Y + +CV
Sbjct: 216 ISLVYTNAPRSFTLIDRGRRNSVLVGRNGFDETYLFSPGSSGIESYS-----KYAY-ICV 377
Query: 279 QPACVEKAITLKPGEEWKG 297
A V + I L P + WKG
Sbjct: 378 GQAAVLQPIVLSPQDVWKG 434
>TC76761 similar to GP|19424061|gb|AAL87257.1 unknown protein {Arabidopsis
thaliana}, partial (52%)
Length = 1356
Score = 28.9 bits (63), Expect = 2.8
Identities = 19/55 (34%), Positives = 28/55 (50%), Gaps = 4/55 (7%)
Frame = +3
Query: 25 NRGFSAEVYL---YGGQVTSWKN-ERGEELLFVSSKPEKEVKKTNQYNCFDMNNN 75
N GF+ +Y YGG S N +GE+ +F SKP K+ N+ + NN+
Sbjct: 399 NGGFNKGIYSNPSYGGNFNSNMNVTKGEDEIFHPSKPSKKNSNPNKKQGENNNNS 563
>TC90850 similar to GP|23496018|gb|AAN35680.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (1%)
Length = 736
Score = 28.1 bits (61), Expect = 4.8
Identities = 22/88 (25%), Positives = 43/88 (48%), Gaps = 7/88 (7%)
Frame = +1
Query: 47 GEELLFVSSKPEKEVKKTNQY--NCFDMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILD 104
GEE L ++S E+ + TN + N F+ N+N+ ++++ L F+ + +W+
Sbjct: 460 GEEFLVMTSNNEQSISSTNSHVNNNFNNNSNQQVRKIMVEHLVDTT---KFIIDSIWM-- 624
Query: 105 PNPPPFPTNSNS-----RAFVDLVLKNS 127
FP +S + R F+ +L+ S
Sbjct: 625 ----NFPVHSTAQVIPLRVFIQEILRRS 696
>TC80483 similar to GP|22136520|gb|AAM91338.1 putative protein {Arabidopsis
thaliana}, partial (32%)
Length = 991
Score = 27.7 bits (60), Expect = 6.2
Identities = 18/51 (35%), Positives = 25/51 (48%)
Frame = +1
Query: 12 MGVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVK 62
+G+N E + GFS +V++ G W NE EEL V K +VK
Sbjct: 724 IGLNIQEAHAFSTSDGFSLDVFVVEG----WPNEETEELKGVLEKENLKVK 864
>BG447553 similar to GP|15292785|gb| putative ribosomal-protein S6 kinase
ATPK19 {Arabidopsis thaliana}, partial (16%)
Length = 665
Score = 27.7 bits (60), Expect = 6.2
Identities = 11/27 (40%), Positives = 18/27 (65%)
Frame = +3
Query: 107 PPPFPTNSNSRAFVDLVLKNSEDDTKN 133
PPP N+NS F+D + NS+++ +N
Sbjct: 189 PPPPSDNNNSLFFLDELTSNSDEEEEN 269
>TC82549 similar to GP|15292785|gb|AAK92761.1 putative ribosomal-protein S6
kinase ATPK19 {Arabidopsis thaliana}, partial (40%)
Length = 705
Score = 27.7 bits (60), Expect = 6.2
Identities = 11/27 (40%), Positives = 18/27 (65%)
Frame = +3
Query: 107 PPPFPTNSNSRAFVDLVLKNSEDDTKN 133
PPP N+NS F+D + NS+++ +N
Sbjct: 60 PPPPSDNNNSLFFLDELTSNSDEEEEN 140
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,532,346
Number of Sequences: 36976
Number of extensions: 151134
Number of successful extensions: 850
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 833
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 838
length of query: 322
length of database: 9,014,727
effective HSP length: 96
effective length of query: 226
effective length of database: 5,465,031
effective search space: 1235097006
effective search space used: 1235097006
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144503.2


