

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144502.3 + phase: 0 /pseudo
(657 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
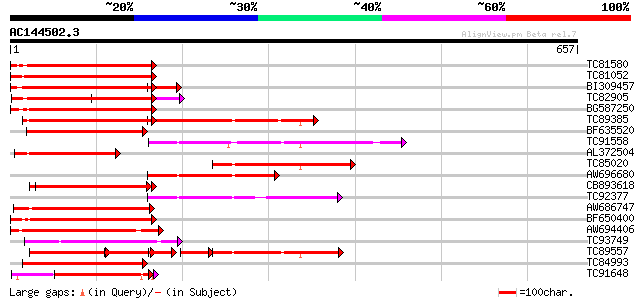
Score E
Sequences producing significant alignments: (bits) Value
TC81580 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Gly... 244 8e-65
TC81052 weakly similar to GP|10178211|dbj|BAB11635. TMV resistan... 231 7e-61
BI309457 weakly similar to GP|15787901|gb resistance gene analog... 230 1e-60
TC82905 weakly similar to GP|3947733|emb|CAA08797.1 NL25 {Solanu... 226 2e-59
BG587250 weakly similar to GP|3947735|emb| NL27 {Solanum tuberos... 225 4e-59
TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 218 6e-57
BF635520 weakly similar to GP|3947735|emb| NL27 {Solanum tuberos... 212 4e-55
TC91558 weakly similar to GP|19774201|gb|AAL99077.1 NBS-LRR-Toll... 199 2e-51
AL372504 weakly similar to GP|7341113|gb| unknown {Glycine max},... 178 7e-45
TC85020 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 174 7e-44
AW696680 similar to GP|19774149|gb NBS-LRR-Toll resistance gene ... 163 2e-40
CB893618 weakly similar to GP|3947733|emb| NL25 {Solanum tuberos... 159 3e-39
TC92377 similar to GP|19774149|gb|AAL99051.1 NBS-LRR-Toll resist... 156 3e-38
AW686747 weakly similar to PIR|A54810|A5 TMV resistance protein ... 154 8e-38
BF650400 weakly similar to GP|9965109|gb| resistance protein MG1... 150 1e-36
AW694406 weakly similar to GP|3947735|emb NL27 {Solanum tuberosu... 148 6e-36
TC93749 similar to GP|3947733|emb|CAA08797.1 NL25 {Solanum tuber... 141 7e-34
TC89557 weakly similar to GP|10178211|dbj|BAB11635. TMV resistan... 106 1e-33
TC84993 weakly similar to GP|3947733|emb|CAA08797.1 NL25 {Solanu... 140 2e-33
TC91648 weakly similar to GP|23477201|emb|CAD36199. NLS-TIR-NBS ... 131 9e-31
>TC81580 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Glycine max},
partial (84%)
Length = 827
Score = 244 bits (623), Expect = 8e-65
Identities = 128/170 (75%), Positives = 136/170 (79%)
Frame = +2
Query: 1 MASSNNSSSSALVTVTLPRRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTN 60
MASSNNSS ALVT + R NYYD F TFRGEDTRNNFT LFDA +GI AFRDDTN
Sbjct: 56 MASSNNSSL-ALVTSS---RNNYYDVFGTFRGEDTRNNFTDFLFDALETKGIFAFRDDTN 223
Query: 61 LPKGESIASELLRAIEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDV 120
L KGESI ELLRAIE S +FVAV SRNYASS WCLQELEKI +CV S+KH+LPVFYDV
Sbjct: 224 LQKGESIEPELLRAIEGSRVFVAVFSRNYASSTWCLQELEKICKCVQRSRKHILPVFYDV 403
Query: 121 DPPVVRKQSGIYCEAFVKHEQIFQQDSQMVLRWREALTQVAGLSGCDLRD 170
DP VVRKQSGIYCEAFVKHEQ FQQD +MV RWREAL V +SG DLRD
Sbjct: 404 DPSVVRKQSGIYCEAFVKHEQRFQQDFEMVSRWREALKHVGSISGWDLRD 553
>TC81052 weakly similar to GP|10178211|dbj|BAB11635. TMV resistance protein
N {Arabidopsis thaliana}, partial (8%)
Length = 656
Score = 231 bits (589), Expect = 7e-61
Identities = 119/170 (70%), Positives = 132/170 (77%)
Frame = +2
Query: 1 MASSNNSSSSALVTVTLPRRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTN 60
MASS+NSSSSAL+ LPRRKNYYD FVTFRGEDTR NF HLF A R+GI AFRDD N
Sbjct: 47 MASSSNSSSSALMA--LPRRKNYYDVFVTFRGEDTRFNFIDHLFAALQRKGIFAFRDDAN 220
Query: 61 LPKGESIASELLRAIEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDV 120
L KGESI EL+RAIE S +F+AVLS+NY+SS WCL+EL IL+C VS + VLPVFYDV
Sbjct: 221 LQKGESIPPELIRAIEGSQVFIAVLSKNYSSSTWCLRELVHILDCSQVSGRRVLPVFYDV 400
Query: 121 DPPVVRKQSGIYCEAFVKHEQIFQQDSQMVLRWREALTQVAGLSGCDLRD 170
DP VR Q GIY EAF KHEQ FQ DS +V WREALTQV +SG DLRD
Sbjct: 401 DPSEVRHQKGIYGEAFSKHEQTFQHDSHVVQSWREALTQVGNISGWDLRD 550
>BI309457 weakly similar to GP|15787901|gb resistance gene analog NBS7
{Helianthus annuus}, partial (45%)
Length = 806
Score = 230 bits (587), Expect = 1e-60
Identities = 119/170 (70%), Positives = 131/170 (77%)
Frame = +3
Query: 1 MASSNNSSSSALVTVTLPRRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTN 60
MAS++NSSS + R+NYYD FVTFRGEDTRNNFT LFDA +GI+ F DDTN
Sbjct: 15 MASTSNSSS----VLGTSSRRNYYDVFVTFRGEDTRNNFTDFLFDALQTKGIIVFSDDTN 182
Query: 61 LPKGESIASELLRAIEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDV 120
LPKGESI ELLRAIE S +FVAV S NYASS WCLQELEKI ECV S KHVLPVFYDV
Sbjct: 183 LPKGESIGPELLRAIEGSQVFVAVFSINYASSTWCLQELEKICECVKGSGKHVLPVFYDV 362
Query: 121 DPPVVRKQSGIYCEAFVKHEQIFQQDSQMVLRWREALTQVAGLSGCDLRD 170
DP VRKQSGIY EAF+KHEQ FQQ+ Q V +WR+AL QV +SG DLRD
Sbjct: 363 DPSDVRKQSGIYGEAFIKHEQRFQQEFQKVSKWRDALKQVGSISGWDLRD 512
Score = 43.1 bits (100), Expect = 3e-04
Identities = 20/40 (50%), Positives = 26/40 (65%)
Frame = +3
Query: 160 VAGLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGP 199
+ G+ L LYD+ISH+F A FIDDV + + LHDGP
Sbjct: 687 MGGIGKTTLAMALYDQISHRFSASWFIDDVKQNY*LHDGP 806
>TC82905 weakly similar to GP|3947733|emb|CAA08797.1 NL25 {Solanum
tuberosum}, partial (22%)
Length = 832
Score = 226 bits (576), Expect = 2e-59
Identities = 116/168 (69%), Positives = 130/168 (77%)
Frame = +1
Query: 3 SSNNSSSSALVTVTLPRRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTNLP 62
+S++ SSSALVT + +KN+YD FVTFRGEDTRNNFT LFDA +GI+ FRD NL
Sbjct: 34 ASSSKSSSALVTSS---KKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQ 204
Query: 63 KGESIASELLRAIEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDVDP 122
KGE I EL RAIE S ++VA+ S+NYASS WCLQELEKI EC+ S KHVLPVFYDVDP
Sbjct: 205 KGECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDP 384
Query: 123 PVVRKQSGIYCEAFVKHEQIFQQDSQMVLRWREALTQVAGLSGCDLRD 170
VRKQSGIY EAFVKHEQ FQQDS V RWREAL QV +SG DLRD
Sbjct: 385 SEVRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRD 528
Score = 52.4 bits (124), Expect = 6e-07
Identities = 37/109 (33%), Positives = 53/109 (47%), Gaps = 2/109 (1%)
Frame = +1
Query: 96 LQELEKILECVH--VSKKHVLPVFYDVDPPVVRKQSGIYCEAFVKHEQIFQQDSQMVLRW 153
+Q++ ILEC + VSK V +D P+ +A H + D +
Sbjct: 559 VQKIINILECKYSCVSKDLV-----GIDSPI---------QALQNHLLLNSVDGVRAI-- 690
Query: 154 REALTQVAGLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGV 202
+ + G+ L LY +ISHQF A CFIDDV+K++ LHD PL V
Sbjct: 691 --GICGMGGIGKTTLATTLYGQISHQFSASCFIDDVTKIYGLHDDPLDV 831
>BG587250 weakly similar to GP|3947735|emb| NL27 {Solanum tuberosum}, partial
(9%)
Length = 655
Score = 225 bits (574), Expect = 4e-59
Identities = 118/170 (69%), Positives = 131/170 (76%)
Frame = +1
Query: 1 MASSNNSSSSALVTVTLPRRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTN 60
MASS+NSS++ V LPRR N YD FVTFRGEDTRNNFT LF A R+GI AFRDDTN
Sbjct: 13 MASSSNSSTAL---VPLPRR-NCYDVFVTFRGEDTRNNFTNFLFAALERKGIYAFRDDTN 180
Query: 61 LPKGESIASELLRAIEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDV 120
LPKGESI ELLR IE S +FVAVLSRNYASS WCLQELEKI EC+ S K+VLP+FY V
Sbjct: 181 LPKGESIGPELLRTIEGSQVFVAVLSRNYASSTWCLQELEKICECIKGSGKYVLPIFYGV 360
Query: 121 DPPVVRKQSGIYCEAFVKHEQIFQQDSQMVLRWREALTQVAGLSGCDLRD 170
DP V+KQSGIY + F KHEQ F+QD V RWREAL QV ++G DLRD
Sbjct: 361 DPSEVKKQSGIYWDDFAKHEQRFKQDPHKVSRWREALNQVGSIAGWDLRD 510
>TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (98%)
Length = 1285
Score = 218 bits (555), Expect = 6e-57
Identities = 111/156 (71%), Positives = 121/156 (77%)
Frame = +3
Query: 15 VTLPRRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRA 74
V LPRR N YD FVTFRGEDTRNNFT LF A R+GI AFRDDTNLPKGESI ELLR
Sbjct: 45 VPLPRR-NCYDVFVTFRGEDTRNNFTNFLFAALERKGIYAFRDDTNLPKGESIGPELLRT 221
Query: 75 IEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDVDPPVVRKQSGIYCE 134
IE S +FVAVLSRNYASS WCLQELEKI EC+ S K+VLP+FY VDP V+KQSGIY +
Sbjct: 222 IEGSQVFVAVLSRNYASSTWCLQELEKICECIKGSGKYVLPIFYGVDPSEVKKQSGIYWD 401
Query: 135 AFVKHEQIFQQDSQMVLRWREALTQVAGLSGCDLRD 170
F KHEQ F+QD V RWREAL QV ++G DLRD
Sbjct: 402 DFAKHEQRFKQDPHKVSRWREALNQVGSIAGWDLRD 509
Score = 190 bits (482), Expect = 2e-48
Identities = 112/204 (54%), Positives = 143/204 (69%), Gaps = 5/204 (2%)
Frame = +3
Query: 160 VAGLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQIC 219
+ G L NLY +I H+F A CFIDDVSK+FRLHDGP+ QKQIL+QT G EH+QIC
Sbjct: 684 MGGKGKTTLAMNLYGQICHRFDASCFIDDVSKIFRLHDGPIDAQKQILHQTLGIEHHQIC 863
Query: 220 NLSTASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGV-CVW*MVRWGSKIIIISRDEHILK 278
N +A++LIR RL R++ LLI DNVD+VEQLE+IGV W + GS+I+IISRDEHILK
Sbjct: 864 NHYSATDLIRHRLSREKTLLILDNVDQVEQLERIGVHREW--LGAGSRIVIISRDEHILK 1037
Query: 279 FFGVDEVYKVPLLDRTNSLQLLCRKAFKLDHILSSMKGW-SMAYYIMLRAHR*QLKYR-- 335
+ VD YKVPLLD T S +L +KAFKL+ I+ MK + ++AY I+ A+ L
Sbjct: 1038EYKVDVDYKVPLLDWTESHKLFRQKAFKLEKII--MKNYQNLAYEILNYANGLPLTITVL 1211
Query: 336 -AHFCGRDIFEWKSALARLRDSPD 358
+ GR++ EWKSALARLR P+
Sbjct: 1212GSFLSGRNVTEWKSALARLRQRPN 1283
>BF635520 weakly similar to GP|3947735|emb| NL27 {Solanum tuberosum}, partial
(8%)
Length = 577
Score = 212 bits (539), Expect = 4e-55
Identities = 103/140 (73%), Positives = 114/140 (80%)
Frame = +1
Query: 20 RKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAIEDSY 79
+KNYYD FVT+RGEDTRNN T LFDA +GI+ FRDD NL KGE I SELLRAIE S+
Sbjct: 76 KKNYYDVFVTYRGEDTRNNLTDFLFDALESKGIMVFRDDINLQKGEYIGSELLRAIERSH 255
Query: 80 IFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDVDPPVVRKQSGIYCEAFVKH 139
++VAV SRNYASS WCLQELEKI EC+ +KHVLP+FYDVDP VRKQSGIY EAFVKH
Sbjct: 256 VYVAVFSRNYASSTWCLQELEKICECIEGLEKHVLPIFYDVDPSEVRKQSGIYWEAFVKH 435
Query: 140 EQIFQQDSQMVLRWREALTQ 159
EQ FQQD QMV RWREAL +
Sbjct: 436 EQRFQQDFQMVSRWREALNK 495
>TC91558 weakly similar to GP|19774201|gb|AAL99077.1 NBS-LRR-Toll resistance
gene analog protein {Medicago edgeworthii}, partial
(60%)
Length = 942
Score = 199 bits (507), Expect = 2e-51
Identities = 123/309 (39%), Positives = 183/309 (58%), Gaps = 10/309 (3%)
Frame = +1
Query: 162 GLSGCDLRDNLYDRISHQ--FGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQIC 219
G+ L + LYD ISHQ F ACCF++DVSK++R G + VQK+IL QT E++ +
Sbjct: 22 GIGKTTLANVLYDTISHQYQFDACCFVEDVSKIYR-DGGAIAVQKRILDQTIKEKNLEGY 198
Query: 220 NLSTASNLIRRRLCRQRVLLIFDNVDKVEQLEK-----IGVCVW*MVRWGSKIIIISRDE 274
+ S S +I RL + ++LL+ DNVD+ QL++ I +C GS+III +RD+
Sbjct: 199 SPSEISGIISNRLYKLKLLLVLDNVDQSVQLQELHINPISLCA------GSRIIITTRDK 360
Query: 275 HILKFFGVDEVYKVPLLDRTNSLQLLCRKAFKLDHILSSMKGWSMAYYIMLRAHR*QLKY 334
HIL + D VY+ LL+ ++ +LLCRKAFK D+ S + + ++ A L
Sbjct: 361 HILIEYEADIVYEAELLNDNDAHELLCRKAFKSDYSSSDYE--ELIPEVLKYAQGLPLAI 534
Query: 335 R---AHFCGRDIFEWKSALARLRDSPDKDVMDVLRLSFDGLEESEKEIFLHIACFFNPSM 391
R + R +W++AL +++PD +M VLR SF+GLE EKEIFLH+ACFF+
Sbjct: 535 RVMGSFLYKRKTAQWRAALEGWQNNPDSGIMKVLRSSFEGLELREKEIFLHVACFFDGER 714
Query: 392 EKYVKNVLNCCGFHADIGLRVLIDKSLISIDESFSSLKEESISMHGLLEELGRKIVQENS 451
E YV+ +L+ CG +IG+ ++KSLI+I + + MH +L+ELG V+E
Sbjct: 715 EDYVRRILHACGLQPNIGIPTSVEKSLITI-------RNQENHMHEMLQELGETNVRETH 873
Query: 452 SKEPRKWSR 460
EPR WS+
Sbjct: 874 PDEPRLWSK 900
>AL372504 weakly similar to GP|7341113|gb| unknown {Glycine max}, partial
(70%)
Length = 478
Score = 178 bits (451), Expect = 7e-45
Identities = 91/123 (73%), Positives = 98/123 (78%)
Frame = +2
Query: 6 NSSSSALVTVTLPRRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTNLPKGE 65
NSS+S+L VT RR NYYD FVTFRGEDTRNNFT +LFDA +GI AFRDDTNL KGE
Sbjct: 113 NSSNSSLALVTSSRR-NYYDVFVTFRGEDTRNNFTDYLFDALETKGIYAFRDDTNLKKGE 289
Query: 66 SIASELLRAIEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDVDPPVV 125
I ELLRAIE S +FVAV SRNYASS WCLQELEKI ECV +KHVLPVFYD+DP V
Sbjct: 290 VIGPELLRAIEGSQVFVAVFSRNYASSTWCLQELEKICECVQGPEKHVLPVFYDIDPSEV 469
Query: 126 RKQ 128
RKQ
Sbjct: 470 RKQ 478
>TC85020 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (55%)
Length = 500
Score = 174 bits (442), Expect = 7e-44
Identities = 101/169 (59%), Positives = 122/169 (71%), Gaps = 4/169 (2%)
Frame = +1
Query: 236 RVLLIFDNVDKVEQLEKIGVC-VW*MVRWGSKIIIISRDEHILKFFGVDEVYKVPLLDRT 294
+ LLIFDNVD+VEQLEKI V W + GS+I+IISRDEHILK +GVD VYKVPL++ T
Sbjct: 1 KALLIFDNVDQVEQLEKIAVHREW--LGAGSRIVIISRDEHILKEYGVDVVYKVPLMNST 174
Query: 295 NSLQLLCRKAFKLDHILSSMKGWSMAYYIMLRAHR*QLKYR---AHFCGRDIFEWKSALA 351
+S +L CRKAFK++ I+ S ++A I+ A L + + G + EWKSALA
Sbjct: 175 DSYELFCRKAFKVEKIIMS-DYQNLANEILDYAKGLPLAIKVLGSFLFGHSVAEWKSALA 351
Query: 352 RLRDSPDKDVMDVLRLSFDGLEESEKEIFLHIACFFNPSMEKYVKNVLN 400
RLR+SP DVMDVL LSFDGL+E EKEIFL IACFFN EKYVKNVLN
Sbjct: 352 RLRESPHNDVMDVLHLSFDGLDEKEKEIFLDIACFFNSQPEKYVKNVLN 498
>AW696680 similar to GP|19774149|gb NBS-LRR-Toll resistance gene analog
protein {Medicago sativa}, partial (96%)
Length = 641
Score = 163 bits (413), Expect = 2e-40
Identities = 93/154 (60%), Positives = 114/154 (73%), Gaps = 1/154 (0%)
Frame = +2
Query: 160 VAGLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQIC 219
+ G+ L LY +ISHQF A CFIDD+SK++R HDG +G QKQIL+QT G E Q+C
Sbjct: 116 MGGIGKTTLATALYGQISHQFDARCFIDDLSKIYR-HDGQVGAQKQILHQTLGVEPFQLC 292
Query: 220 NLSTASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGV-CVW*MVRWGSKIIIISRDEHILK 278
NL ++L+RRRL R RVL+I DNVDKV QL+K+GV W + GS+IIIIS DEHILK
Sbjct: 293 NLYHTTDLMRRRLRRLRVLIIVDNVDKVGQLDKLGVNREW--LGAGSRIIIISGDEHILK 466
Query: 279 FFGVDEVYKVPLLDRTNSLQLLCRKAFKLDHILS 312
+GVD VY+VPLL+ TNSLQL KAFKL HI+S
Sbjct: 467 EYGVDVVYRVPLLNWTNSLQLFSLKAFKLYHIIS 568
>CB893618 weakly similar to GP|3947733|emb| NL25 {Solanum tuberosum}, partial
(18%)
Length = 927
Score = 159 bits (402), Expect = 3e-39
Identities = 85/141 (60%), Positives = 97/141 (68%)
Frame = +2
Query: 30 FRGEDTRNNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAIEDSYIFVAVLSRNY 89
FRG DTR NFT HLF A R+ I FRDDTNL KG SIAS+L++AIE S +F+ V S+NY
Sbjct: 2 FRGTDTRFNFTDHLFGALQRKRIFTFRDDTNLQKGNSIASDLIQAIEGSQVFIVVFSKNY 181
Query: 90 ASSIWCLQELEKILECVHVSKKHVLPVFYDVDPPVVRKQSGIYCEAFVKHEQIFQQDSQM 149
ASS WCL+EL IL C + K VLPVFYDVD VRKQSG Y E+F H + FQ S M
Sbjct: 182 ASSTWCLRELAYILNCSVLYGKRVLPVFYDVDLSEVRKQSGGYGESFNYHGKRFQDHSNM 361
Query: 150 VLRWREALTQVAGLSGCDLRD 170
V R RE L V +SG DLRD
Sbjct: 362 VQRRRETLQLVGNISGWDLRD 424
Score = 156 bits (395), Expect = 2e-38
Identities = 84/141 (59%), Positives = 93/141 (65%)
Frame = +3
Query: 24 YDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAIEDSYIFVA 83
YD FV+FRG DTR NFT H A R GI AFRDDT L KGE IA L RAIE S +++
Sbjct: 519 YDVFVSFRGPDTRFNFTDHFCAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQVYIV 698
Query: 84 VLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDVDPPVVRKQSGIYCEAFVKHEQIF 143
V S+NYASS WCL+ELE IL C KHVLPVFYDVDP V+KQSG Y +A KH
Sbjct: 699 VFSKNYASSTWCLRELEYILHCSKKHGKHVLPVFYDVDPSEVQKQSGGYGDALSKH---- 866
Query: 144 QQDSQMVLRWREALTQVAGLS 164
MV RWRE LTQV +S
Sbjct: 867 --GLNMV*RWRETLTQVGNIS 923
>TC92377 similar to GP|19774149|gb|AAL99051.1 NBS-LRR-Toll resistance gene
analog protein {Medicago sativa}, partial (57%)
Length = 945
Score = 156 bits (394), Expect = 3e-38
Identities = 97/226 (42%), Positives = 133/226 (57%)
Frame = +1
Query: 160 VAGLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQIC 219
+ G+ L LYDRISHQF ACCFIDD+SK++R ++G L ++ + + +
Sbjct: 280 MGGIGKTTLATALYDRISHQFDACCFIDDISKVYR-NEGRLVLKSKHYIKLLANSTFRYA 456
Query: 220 NLSTASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGVCVW*MVRWGSKIIIISRDEHILKF 279
+T +NLIR +L R LL+ DNVD+VEQ EK+ V + GS+I+I SRD HILK
Sbjct: 457 IFTTKTNLIRLKLGHIRALLVLDNVDQVEQQEKLAVSSE-CLGAGSRIVITSRDVHILKE 633
Query: 280 FGVDEVYKVPLLDRTNSLQLLCRKAFKLDHILSSMKGWSMAYYIMLRAHR*QLKYRAHFC 339
+ VDE+ + C+KAF+ I S+ + + + C
Sbjct: 634 YEVDEI-----------VN*FCQKAFQGGKIKSNYEELTCHIKLC*WPTTGN*SIGLILC 780
Query: 340 GRDIFEWKSALARLRDSPDKDVMDVLRLSFDGLEESEKEIFLHIAC 385
GR+I EW SALARLR++P+KD+MDVLRLSF+GLEESE IFL I C
Sbjct: 781 GRNISEWTSALARLRENPNKDIMDVLRLSFEGLEESENVIFLDIVC 918
>AW686747 weakly similar to PIR|A54810|A5 TMV resistance protein N - tobacco
(Nicotiana glutinosa), partial (10%)
Length = 644
Score = 154 bits (390), Expect = 8e-38
Identities = 81/163 (49%), Positives = 108/163 (65%)
Frame = +3
Query: 5 NNSSSSALVTVTLPRRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTNLPKG 64
+ +SSS+ ++ R N+ F++FRGEDTR FT HLF + R GI F+DD +L +G
Sbjct: 69 DEASSSSSPSIQTSRWTNH--VFLSFRGEDTRQGFTDHLFASLERRGIKTFKDDHDLERG 242
Query: 65 ESIASELLRAIEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDVDPPV 124
E I+ EL +AIE+S + +LS NYASS WCL EL+KI+EC + V P+FY VDP
Sbjct: 243 EVISYELNKAIEESMFAIIILSPNYASSTWCLDELKKIVECSKSFGQAVFPIFYGVDPSD 422
Query: 125 VRKQSGIYCEAFVKHEQIFQQDSQMVLRWREALTQVAGLSGCD 167
VR Q G + EAF KHE+ F++D V RWR+AL +VAG SG D
Sbjct: 423 VRHQRGSFDEAFRKHEEKFRKDRTKVERWRDALREVAGYSGWD 551
>BF650400 weakly similar to GP|9965109|gb| resistance protein MG13 {Glycine
max}, partial (36%)
Length = 644
Score = 150 bits (379), Expect = 1e-36
Identities = 74/170 (43%), Positives = 115/170 (67%)
Frame = +1
Query: 1 MASSNNSSSSALVTVTLPRRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTN 60
M+S+ +S SS+ V P++K YD F++FRG+DTR NFT HL+D +R+ + F D+
Sbjct: 97 MSSAFSSVSSSSVA---PQKK--YDVFLSFRGDDTRRNFTSHLYDTLSRKKVETFIDNNE 261
Query: 61 LPKGESIASELLRAIEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDV 120
L KG+ I+ L++AIE+S++ + + S NYASS WCL EL+KIL+C ++ V+PVFY++
Sbjct: 262 LEKGDEISPALIKAIEESHVSIIIFSENYASSKWCLNELKKILQCKKYMQQIVIPVFYNI 441
Query: 121 DPPVVRKQSGIYCEAFVKHEQIFQQDSQMVLRWREALTQVAGLSGCDLRD 170
DP VRKQ+G Y +AF KH + + ++ + W+ AL + A L G D ++
Sbjct: 442 DPSHVRKQTGSYEQAFTKHMRDLKLNNDKLQEWKAALAEAASLVGWDFQN 591
>AW694406 weakly similar to GP|3947735|emb NL27 {Solanum tuberosum}, partial
(14%)
Length = 628
Score = 148 bits (374), Expect = 6e-36
Identities = 76/178 (42%), Positives = 112/178 (62%)
Frame = +3
Query: 1 MASSNNSSSSALVTVTLPRRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTN 60
MA N +S+ + TL N +D F++FRG+DTR FT HL +A + G+ F DD+
Sbjct: 48 MAMQINIGASS--SSTLEVASNSFDVFISFRGDDTRRKFTSHLNEALKKSGVKTFIDDSE 221
Query: 61 LPKGESIASELLRAIEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDV 120
L KG+ I+S L++AIE+S + + S +YASS WCL EL KILEC + + V+P+FY++
Sbjct: 222 LKKGDEISSALIKAIEESCASIVIFSEDYASSKWCLNELVKILECKKDNGQIVIPIFYEI 401
Query: 121 DPPVVRKQSGIYCEAFVKHEQIFQQDSQMVLRWREALTQVAGLSGCDLRDNLYDRISH 178
DP VR Q G Y +AF KHE+ +Q +W++ALT+V+ LSG D + + + H
Sbjct: 402 DPSHVRNQIGSYGQAFAKHEKNLKQQ-----KWKDALTEVSNLSGWDSKSSRIESYFH 560
>TC93749 similar to GP|3947733|emb|CAA08797.1 NL25 {Solanum tuberosum},
partial (17%)
Length = 772
Score = 141 bits (356), Expect = 7e-34
Identities = 76/183 (41%), Positives = 109/183 (59%)
Frame = +2
Query: 18 PRRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAIED 77
P + + F++FRGEDTR FT HL R + + D NL +G+ I+S LL AIE+
Sbjct: 32 PHHQKHGQVFLSFRGEDTRYTFTSHLHATLTRLKVGTYID-YNLQRGDEISSTLLMAIEE 208
Query: 78 SYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDVDPPVVRKQSGIYCEAFV 137
+ + + + S+NY +S WCL EL KILEC + + +LP+FYD+DP VR Q+G Y EAFV
Sbjct: 209 AKVSIVIFSKNYGNSKWCLDELVKILECKKMKGQILLPIFYDIDPSHVRNQTGSYAEAFV 388
Query: 138 KHEQIFQQDSQMVLRWREALTQVAGLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHD 197
KHE+ FQ + V WR AL + A +SG + N R+ + D + K+ R++D
Sbjct: 389 KHEKQFQGKLEKVQTWRHALREAANISGWECSVN---RMESELLEKIAKDVIEKLNRVND 559
Query: 198 GPL 200
G L
Sbjct: 560 GDL 568
>TC89557 weakly similar to GP|10178211|dbj|BAB11635. TMV resistance protein N
{Arabidopsis thaliana}, partial (8%)
Length = 1567
Score = 106 bits (265), Expect(3) = 7e-33
Identities = 59/155 (38%), Positives = 98/155 (63%), Gaps = 3/155 (1%)
Frame = +3
Query: 236 RVLLIFDNVDKVEQLEKIGVCVW*MVRWGSKIIIISRDEHILKFFGVDEVYKVPLLDRTN 295
++L++ DNV+++EQLEK+ + + SKIIII+RD+HIL+ +G DEV++ L++ +
Sbjct: 1110 KLLIVLDNVEQLEQLEKLDIEPK-FLHPRSKIIIITRDKHILQAYGADEVFEAKLMNDED 1286
Query: 296 SLQLLCRKAFKLDHILSSMKGWSMAYYIMLRAHR*QLKYR---AHFCGRDIFEWKSALAR 352
+ +LLCRKAFK D+ S + +++ A R L + + R+ EW S L +
Sbjct: 1287 AHKLLCRKAFKSDYPSSGFA--ELIPKVLVYAQRLPLAVKVLGSFLFSRNANEWSSTLDK 1460
Query: 353 LRDSPDKDVMDVLRLSFDGLEESEKEIFLHIACFF 387
+P +M L++S++GLE+ EKE+FLH+A FF
Sbjct: 1461 FEKNPPNKIMKALQVSYEGLEKDEKEVFLHVAXFF 1565
Score = 95.5 bits (236), Expect(2) = 1e-33
Identities = 46/94 (48%), Positives = 61/94 (63%)
Frame = +3
Query: 24 YDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAIEDSYIFVA 83
YD F++FRG DTRN+F HL+ NR+GI F+DD L KGE+I+ +LL+AI+ S I +
Sbjct: 264 YDVFISFRGSDTRNSFVDHLYAHLNRKGIFTFKDDKQLQKGEAISPQLLQAIQQSRISIV 443
Query: 84 VLSRNYASSIWCLQELEKILECVHVSKKHVLPVF 117
V S++YASS WCL E+ I E K P F
Sbjct: 444 VFSKDYASSTWCLDEMAAINESRINLKTSCFPCF 545
Score = 66.2 bits (160), Expect(2) = 1e-33
Identities = 30/60 (50%), Positives = 41/60 (68%)
Frame = +1
Query: 110 KKHVLPVFYDVDPPVVRKQSGIYCEAFVKHEQIFQQDSQMVLRWREALTQVAGLSGCDLR 169
K+ V PVFYDVDP V+KQ+G+Y AFV H + F+ +S V RWR +T + G +G D+R
Sbjct: 523 KQVVFPVFYDVDPSHVKKQNGVYENAFVLHTEAFKHNSGKVARWRTTMTYLGGTAGWDVR 702
Score = 39.7 bits (91), Expect(3) = 7e-33
Identities = 17/32 (53%), Positives = 23/32 (71%)
Frame = +1
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMF 193
G+ L LYD+IS QF ACCFI++VSK++
Sbjct: 889 GIGKTTLATVLYDKISFQFDACCFIENVSKIY 984
Score = 33.5 bits (75), Expect(3) = 7e-33
Identities = 15/38 (39%), Positives = 23/38 (60%)
Frame = +2
Query: 199 PLGVQKQILYQTHGEEHNQICNLSTASNLIRRRLCRQR 236
P+ VQKQI+ QT E++ C+ S +R RLC+ +
Sbjct: 998 PVAVQKQIICQTIEEKNIDTCSARKISQTMRNRLCKTK 1111
>TC84993 weakly similar to GP|3947733|emb|CAA08797.1 NL25 {Solanum
tuberosum}, partial (21%)
Length = 672
Score = 140 bits (352), Expect = 2e-33
Identities = 66/144 (45%), Positives = 95/144 (65%)
Frame = +2
Query: 16 TLPRRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAI 75
TL N +D F++FRG+DTR FT HL +A + G+ F DD L KG+ I+S L++AI
Sbjct: 92 TLEVASNSFDVFISFRGDDTRRKFTSHLNEALKKSGLKTFIDDNELKKGDEISSALIKAI 271
Query: 76 EDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDVDPPVVRKQSGIYCEA 135
E+S + +LS NYASS WCL EL KILEC + + V+P+FY++DP VR Q G Y +A
Sbjct: 272 EESCASIVILSENYASSKWCLNELVKILECKKDNGQIVIPIFYEIDPSHVRYQIGSYGQA 451
Query: 136 FVKHEQIFQQDSQMVLRWREALTQ 159
F K+E+ + + +W++ALT+
Sbjct: 452 FAKYEKNLRHKKDNLQKWKDALTE 523
>TC91648 weakly similar to GP|23477201|emb|CAD36199. NLS-TIR-NBS disease
resistance protein {Populus tremula}, partial (17%)
Length = 666
Score = 131 bits (329), Expect = 9e-31
Identities = 71/118 (60%), Positives = 82/118 (69%), Gaps = 3/118 (2%)
Frame = +2
Query: 52 ILAFRDDTNLPKGESIASELLRAIEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKK 111
I +FRDDT L KGESIA ELLRAIEDS IFV V S+NYASS+WCL+ELE IL+ +S K
Sbjct: 254 IFSFRDDTKLKKGESIAPELLRAIEDSQIFVVVFSKNYASSVWCLRELECILQSFQLSGK 433
Query: 112 HVLPVFYDVDPPVVRKQSGIYCEAFVKHEQIFQQDSQMVLRWREALTQVAGLS---GC 166
VLPVFYDVDP VR Q G Y EA KHE+ FQQ+ + + E T +G S GC
Sbjct: 434 RVLPVFYDVDPSEVRYQKGCYAEALAKHEERFQQNFENXAK-MEGSTDTSGQSLWMGC 604
Score = 63.5 bits (153), Expect = 2e-10
Identities = 56/178 (31%), Positives = 85/178 (47%), Gaps = 8/178 (4%)
Frame = +3
Query: 3 SSNNS----SSSALVTVTLPRRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDD 58
SSNNS SSS+ +++ R+NYYD FV+FRG+DTR NFT HLF A R+ +F
Sbjct: 96 SSNNSIIKISSSSSSSISSSPRRNYYDVFVSFRGKDTRLNFTDHLFAALQRK---SFFHS 266
Query: 59 TNLPKGESIASELLRAIEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFY 118
+ + L + E +L + + + E + ++ VF
Sbjct: 267 GMILS*RKVNP*HLNSFEQLKTLRFLLWFSLRTMLPQFGVCEN-WSAYFKAFNYLENVFC 443
Query: 119 DVDPPVVRKQSGIYCEAFVKHEQIFQQDSQMVL----RWREALTQVAGLSGCDLRDNL 172
++ ++ I + +K ++DS L RWREALTQVA LSG D+R +
Sbjct: 444 LCSMMLIHQRCDIKKDVMLKP*LNMKKDSNKTLKIXQRWREALTQVANLSGWDVRXKI 617
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.333 0.145 0.451
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,407,329
Number of Sequences: 36976
Number of extensions: 337348
Number of successful extensions: 2470
Number of sequences better than 10.0: 209
Number of HSP's better than 10.0 without gapping: 2294
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2343
length of query: 657
length of database: 9,014,727
effective HSP length: 102
effective length of query: 555
effective length of database: 5,243,175
effective search space: 2909962125
effective search space used: 2909962125
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144502.3


