

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144478.12 + phase: 0 /pseudo
(1250 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
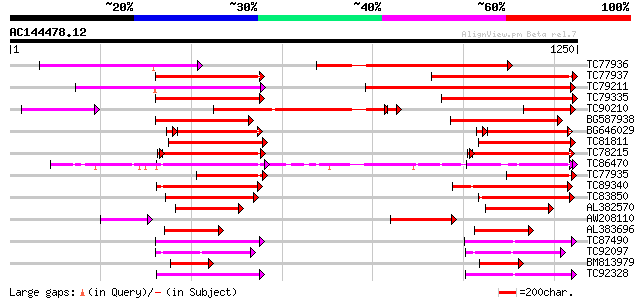
Score E
Sequences producing significant alignments: (bits) Value
TC77936 weakly similar to PIR|T52319|T52319 P-glycoprotein-like ... 506 e-143
TC77937 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japo... 433 e-121
TC79211 similar to GP|4204793|gb|AAD10836.1| P-glycoprotein {Sol... 428 e-120
TC79335 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japo... 418 e-117
TC90210 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japo... 390 e-113
BG587938 homologue to GP|22655312|g putative ABC transporter {Ar... 313 2e-85
BG646029 similar to GP|14715462|d CjMDR1 {Coptis japonica}, part... 261 1e-74
TC81811 similar to GP|6671365|gb|AAF23176.1| P-glycoprotein {Gos... 276 5e-74
TC78215 similar to GP|10177549|dbj|BAB10828. ABC transporter-lik... 232 1e-60
TC86470 similar to PIR|F84919|F84919 glutathione-conjugate trans... 222 8e-58
TC77935 similar to PIR|E85023|E85023 probable P-glycoprotein-lik... 220 2e-57
TC89340 weakly similar to GP|17427488|emb|CAD14006. PROBABLE COM... 205 1e-52
TC83850 weakly similar to GP|7300071|gb|AAF55241.1| CG4225 gene ... 202 9e-52
AL382570 weakly similar to GP|10241621|em putative P-glycoprotei... 181 2e-45
AW208110 similar to GP|14715462|d CjMDR1 {Coptis japonica}, part... 175 9e-44
AL383696 homologue to PIR|T47671|T4 P-glycoprotein-like - Arabid... 147 3e-35
TC87490 similar to PIR|T52080|T52080 multi resistance protein [i... 145 1e-34
TC92097 similar to GP|22553016|emb|CAD44995. multidrug-resistanc... 145 1e-34
BM813979 homologue to PIR|E85023|E85 probable P-glycoprotein-lik... 144 2e-34
TC92328 similar to PIR|T52080|T52080 multi resistance protein [i... 136 6e-32
>TC77936 weakly similar to PIR|T52319|T52319 P-glycoprotein-like protein pgp3
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1219
Score = 506 bits (1302), Expect = e-143
Identities = 251/432 (58%), Positives = 324/432 (74%)
Frame = +1
Query: 677 GFLFSAVISMFYKPPEQQRKESRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRS 736
G L S +I++FYKP + R +S+ W+++FV + + TL+I+P + +FFG+AGGKLI+RIR+
Sbjct: 1 GLLISKMINIFYKPAHELRHDSKVWAIVFVAVAVATLLIIPCRFYFFGVAGGKLIQRIRN 180
Query: 737 LTFEKIVHQEISWFDDPSHSRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDAS 796
+ FEK+VH E+SWFD+ HS SGA+GARLS DA+
Sbjct: 181 MCFEKVVHMEVSWFDEAEHS---------------------------SGALGARLSTDAA 279
Query: 797 TVKSLVGDTMALIVQNISTVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFS 856
+V++LVGD + L+VQNI+T IAGLVI+F A+W LAFIVL L P++ + G VQ+K LKGFS
Sbjct: 280 SVRALVGDALGLLVQNIATAIAGLVISFQASWQLAFIVLALAPLLGLNGYVQVKVLKGFS 459
Query: 857 ADAKVMYEEASQVANDAVSSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFG 916
ADAK +YEEASQVANDAV SIRTVASFCAE KVM++Y +KC GP K+GVR G++SG GFG
Sbjct: 460 ADAKKLYEEASQVANDAVGSIRTVASFCAEKKVMELYKQKCEGPIKKGVRRGIISGFGFG 639
Query: 917 CSFLVLYCTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKD 976
SF +LY +A +FY G+ LV+ GK+TF++VF VFFALTM A+ VSQ+ TL PD+ AK
Sbjct: 640 LSFFMLYAVDACVFYAGARLVEDGKSTFSDVFLVFFALTMAAMGVSQSGTLVPDSTNAKS 819
Query: 977 SAASIFEIIDSKPDIDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSA 1036
+AASIF I+D IDSS +G+T E V GDIE HV+F YPT D+QIF DL L+I S
Sbjct: 820 AAASIFAILDHNSQIDSSDESGMTLEEVKGDIEFNHVSFKYPTTLDVQIFNDLCLNIRSG 999
Query: 1037 KTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILF 1096
KT+ALVGESGSGKSTVISLL+RFYDP+SG I LDG++++ ++ WLRQQMGLV QEPILF
Sbjct: 1000 KTVALVGESGSGKSTVISLLQRFYDPDSGHITLDGIEIQRMQVKWLRQQMGLVSQEPILF 1179
Query: 1097 NESIRANIGYGK 1108
N+++ ANI Y K
Sbjct: 1180 NDTVXANIAYXK 1215
Score = 230 bits (586), Expect = 3e-60
Identities = 139/406 (34%), Positives = 216/406 (52%), Gaps = 46/406 (11%)
Frame = +1
Query: 66 LFLGNVINAFGSSNPADAIKQVSKV-SLLFVYLAIGSGIASFLQVTCWMVTGERQAARIR 124
L + +IN F PA ++ SKV +++FV +A+ + + + + V G + RIR
Sbjct: 4 LLISKMINIF--YKPAHELRHDSKVWAIVFVAVAVATLLIIPCRFYFFGVAGGKLIQRIR 177
Query: 125 SLYLKTILQQDIAFFD-TETNTGEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVM 183
++ + ++ ++++FD E ++G + R+S D ++ +G+ +G Q + G V+
Sbjct: 178 NMCFEKVVHMEVSWFDEAEHSSGALGARLSTDAASVRALVGDALGLLVQNIATAIAGLVI 357
Query: 184 AFIKGWRLAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVAS 243
+F W+LA ++LA P + + G V+ S+ + Y EA V + VG+IRTVAS
Sbjct: 358 SFQASWQLAFIVLALAPLLGLNGYVQVKVLKGFSADAKKLYEEASQVANDAVGSIRTVAS 537
Query: 244 FTGEKKAIEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGY 303
F EKK +E Y K + V++GI+SGFG G+ F+ + + G++LV +
Sbjct: 538 FCAEKKVMELYKQKCEGPIKKGVRRGIISGFGFGLSFFMLYAVDACVFYAGARLVEDGKS 717
Query: 304 NGGTVMTVIIAL--------------------------------------------MTGG 319
V V AL MT
Sbjct: 718 TFSDVFLVFFALTMAAMGVSQSGTLVPDSTNAKSAAASIFAILDHNSQIDSSDESGMTLE 897
Query: 320 IIKGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDP 379
+KGDIE VSF+YP DVQIF+ L + SG T ALVG+SGSGKSTVISLL+RFYDP
Sbjct: 898 EVKGDIEFNHVSFKYPTTLDVQIFNDLCLNIRSGKTVALVGESGSGKSTVISLLQRFYDP 1077
Query: 380 DAGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGK 425
D+G + +DG+ ++ +Q++W+R+Q+GLVSQEPILF ++ NIAY K
Sbjct: 1078DSGHITLDGIEIQRMQVKWLRQQMGLVSQEPILFNDTVXANIAYXK 1215
>TC77937 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japonica},
partial (24%)
Length = 1164
Score = 433 bits (1113), Expect = e-121
Identities = 217/321 (67%), Positives = 268/321 (82%)
Frame = +3
Query: 930 FYIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKP 989
FY G+ LV+ GK++F++VFRVFFAL+M AI +SQ+ +L PD+ KAK + ASIF I+D K
Sbjct: 15 FYAGARLVEDGKSSFSDVFRVFFALSMAAIGLSQSGSLLPDSTKAKSAVASIFAILDRKS 194
Query: 990 DIDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGK 1049
ID + +G+T E V G+IE +HVNF YPTRPDIQIF+DL L+I S KT+ALVGESGSGK
Sbjct: 195 LIDPTDESGITLEEVKGEIEFKHVNFKYPTRPDIQIFRDLCLNIHSGKTVALVGESGSGK 374
Query: 1050 STVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKE 1109
STVISL++RFYDP+SG I LDG ++++ ++ WLRQQMGLV QEP+LFN++IRANI YGK
Sbjct: 375 STVISLIQRFYDPDSGHITLDGKEIQSLQVKWLRQQMGLVSQEPVLFNDTIRANIAYGKG 554
Query: 1110 GGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKIL 1169
G A+E EIIAAA ANAH FIS+L GYDT VGERG QLSGGQKQR+AIAR ++KNPKIL
Sbjct: 555 GDASEAEIIAAAELANAHKFISSLQKGYDTVVGERGVQLSGGQKQRVAIARAIVKNPKIL 734
Query: 1170 LLDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRH 1229
LLDEATSALDAESE++VQ+ALDRV V RTT++VAHRL+TI+GAD IAV+KNG +AEKG+H
Sbjct: 735 LLDEATSALDAESEKVVQDALDRVMVERTTIIVAHRLSTIKGADLIAVVKNGVIAEKGKH 914
Query: 1230 DELMRITDGVYASLVALHSSA 1250
+ L+ G YASLVALH+SA
Sbjct: 915 EALLH-KGGDYASLVALHTSA 974
Score = 300 bits (769), Expect = 2e-81
Identities = 147/243 (60%), Positives = 196/243 (80%), Gaps = 1/243 (0%)
Frame = +3
Query: 321 IKGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPD 380
+KG+IE + V+F+YP RPD+QIF L + SG T ALVG+SGSGKSTVISL++RFYDPD
Sbjct: 237 VKGEIEFKHVNFKYPTRPDIQIFRDLCLNIHSGKTVALVGESGSGKSTVISLIQRFYDPD 416
Query: 381 AGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEG-ATDEEITTAITL 439
+G + +DG +++LQ++W+R+Q+GLVSQEP+LF +IR NIAYGK G A++ EI A L
Sbjct: 417 SGHITLDGKEIQSLQVKWLRQQMGLVSQEPVLFNDTIRANIAYGKGGDASEAEIIAAAEL 596
Query: 440 ANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESE 499
ANA KFI L +G DT+ G+ G QLSGGQKQR+AIARAI+KNPKILLLDEATSALDAESE
Sbjct: 597 ANAHKFISSLQKGYDTVVGERGVQLSGGQKQRVAIARAIVKNPKILLLDEATSALDAESE 776
Query: 500 RIVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQL 559
++VQ+AL++++++RTT++VAHRL+TI+ AD+IAVV+ G I E+G H L + G Y+ L
Sbjct: 777 KVVQDALDRVMVERTTIIVAHRLSTIKGADLIAVVKNGVIAEKGKHEAL-LHKGGDYASL 953
Query: 560 IRL 562
+ L
Sbjct: 954 VAL 962
>TC79211 similar to GP|4204793|gb|AAD10836.1| P-glycoprotein {Solanum
tuberosum}, partial (36%)
Length = 1667
Score = 428 bits (1101), Expect = e-120
Identities = 217/465 (46%), Positives = 325/465 (69%), Gaps = 2/465 (0%)
Frame = +1
Query: 784 SGAVGARLSIDASTVKSLVGDTMALIVQNISTVIAGLVIAFTANWILAFIVLVLTPMILM 843
S + ARL++DA+ V+S +GD +++IVQN + ++ F W LA +++ + P+++
Sbjct: 7 SARISARLALDANNVRSAIGDRISVIVQNTALMLVACTAGFVLQWRLALVLIAVFPVVVA 186
Query: 844 QGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIRTVASFCAESKVMDMYSKKCLGPAKQ 903
++Q F+ GFS D + + +A+Q+A +A++++RTVA+F +ESK++ +++ P ++
Sbjct: 187 ATVLQKMFMTGFSGDLEAAHAKATQLAGEAIANVRTVAAFNSESKIVRLFASNLETPLQR 366
Query: 904 GVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQ 963
G +SG G+G + LY + A + S LV+HG + F++ RVF L ++A ++
Sbjct: 367 CFWKGQISGSGYGIAQFALYASYALGLWYASWLVKHGISDFSKTIRVFMVLMVSANGAAE 546
Query: 964 TTTLAPDTNKAKDSAASIFEIIDSKPDID-SSSNAGVTRETVVGDIELQHVNFNYPTRPD 1022
T TLAPD K + S+F+++D + +I+ +A + + G++EL+HV+F+YPTRPD
Sbjct: 547 TLTLAPDFIKGGRAMRSVFDLLDRQTEIEPDDQDATPVPDRLRGEVELKHVDFSYPTRPD 726
Query: 1023 IQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWL 1082
+ +F+DL+L I + KT+ALVG SG GKS+VI+L++RFYDP SGRI++DG D++ + L L
Sbjct: 727 MPVFRDLNLRIRAGKTLALVGPSGCGKSSVIALIQRFYDPTSGRIMIDGKDIRKYNLKSL 906
Query: 1083 RQQMGLVGQEPILFNESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVG 1142
R+ + +V QEP LF +I NI YG + ATE EII AA ANAH FIS+LPDGY T VG
Sbjct: 907 RRHISVVPQEPCLFATTIYENIAYGHDS-ATEAEIIEAATLANAHKFISSLPDGYKTFVG 1083
Query: 1143 ERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRTTVVV 1202
ERG QLSGGQKQRIA+AR L+ +++LLDEATSALDAESER VQEALDR S +TT++V
Sbjct: 1084 ERGVQLSGGQKQRIAVARAFLRKAELMLLDEATSALDAESERSVQEALDRASTGKTTIIV 1263
Query: 1203 AHRLTTIRGADTIAVIKNGAVAEKGRHDELMR-ITDGVYASLVAL 1246
AHRL+TIR A+ IAVI +G VAE+G H +LM+ DG+YA ++ L
Sbjct: 1264 AHRLSTIRNANVIAVIDDGKVAEQGSHSQLMKNHQDGIYARMIQL 1398
Score = 327 bits (838), Expect = 2e-89
Identities = 181/465 (38%), Positives = 266/465 (56%), Gaps = 46/465 (9%)
Frame = +1
Query: 145 TGEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVAV 204
+ + R++ D ++ A+G+++ Q + F+ WRLA+VL+A P V
Sbjct: 7 SARISARLALDANNVRSAIGDRISVIVQNTALMLVACTAGFVLQWRLALVLIAVFPVVVA 186
Query: 205 AGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYTT 264
A + M S + A+A+A + + + +RTVA+F E K + + S ++
Sbjct: 187 ATVLQKMFMTGFSGDLEAAHAKATQLAGEAIANVRTVAAFNSESKIVRLFASNLETPLQR 366
Query: 265 MVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMT------- 317
+G +SG G G+ F + +Y L +WY S LV + + V + LM
Sbjct: 367 CFWKGQISGSGYGIAQFALYASYALGLWYASWLVKHGISDFSKTIRVFMVLMVSANGAAE 546
Query: 318 ----------GGI----------------------------IKGDIELRDVSFRYPARPD 339
GG ++G++EL+ V F YP RPD
Sbjct: 547 TLTLAPDFIKGGRAMRSVFDLLDRQTEIEPDDQDATPVPDRLRGEVELKHVDFSYPTRPD 726
Query: 340 VQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWI 399
+ +F +L + +G T ALVG SG GKS+VI+L++RFYDP +G ++IDG +++ L+ +
Sbjct: 727 MPVFRDLNLRIRAGKTLALVGPSGCGKSSVIALIQRFYDPTSGRIMIDGKDIRKYNLKSL 906
Query: 400 REQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQ 459
R I +V QEP LF T+I ENIAYG + AT+ EI A TLANA KFI LP G T G+
Sbjct: 907 RRHISVVPQEPCLFATTIYENIAYGHDSATEAEIIEAATLANAHKFISSLPDGYKTFVGE 1086
Query: 460 NGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVA 519
G QLSGGQKQRIA+ARA L+ +++LLDEATSALDAESER VQEAL++ +TT++VA
Sbjct: 1087RGVQLSGGQKQRIAVARAFLRKAELMLLDEATSALDAESERSVQEALDRASTGKTTIIVA 1266
Query: 520 HRLTTIRNADIIAVVQQGKIVERGTHSGLTMD-PDGAYSQLIRLQ 563
HRL+TIRNA++IAV+ GK+ E+G+HS L + DG Y+++I+LQ
Sbjct: 1267HRLSTIRNANVIAVIDDGKVAEQGSHSQLMKNHQDGIYARMIQLQ 1401
>TC79335 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japonica},
partial (23%)
Length = 1135
Score = 418 bits (1075), Expect = e-117
Identities = 206/299 (68%), Positives = 253/299 (83%)
Frame = +2
Query: 952 FALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTRETVVGDIELQ 1011
FALTM AI +SQ+++ APD++KAK + ASIF +ID K ID S +G T +++ G+IEL+
Sbjct: 2 FALTMAAIGISQSSSFAPDSSKAKSATASIFGMIDKKSKIDPSEESGTTLDSIKGEIELR 181
Query: 1012 HVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDG 1071
H++F YP+RPDIQIF+DL+L+I S KT+ALVGESGSGKSTVI+LL+RFYDP+SG I LDG
Sbjct: 182 HISFKYPSRPDIQIFRDLNLTIHSGKTVALVGESGSGKSTVIALLQRFYDPDSGEITLDG 361
Query: 1072 VDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGATEDEIIAAANAANAHSFIS 1131
++++ +L WLRQQMGLV QEP+LFN++IRANI YGK G ATE EIIAAA ANAH FIS
Sbjct: 362 IEIRQLQLKWLRQQMGLVSQEPVLFNDTIRANIAYGKGGIATEAEIIAAAELANAHRFIS 541
Query: 1132 NLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESERIVQEALD 1191
L GYDT VGERGTQLSGGQKQR+AIAR ++K+PKILLLDEATSALDAESER+VQ+ALD
Sbjct: 542 GLQQGYDTIVGERGTQLSGGQKQRVAIARAIIKSPKILLLDEATSALDAESERVVQDALD 721
Query: 1192 RVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELMRITDGVYASLVALHSSA 1250
+V VNRTTVVVAHRL+TI+ AD IAV+KNG + EKGRH+ L+ + DG YASLV LH+SA
Sbjct: 722 KVMVNRTTVVVAHRLSTIKNADVIAVVKNGVIVEKGRHETLINVKDGFYASLVQLHTSA 898
Score = 323 bits (829), Expect = 2e-88
Identities = 160/243 (65%), Positives = 203/243 (82%), Gaps = 1/243 (0%)
Frame = +2
Query: 321 IKGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPD 380
IKG+IELR +SF+YP+RPD+QIF +L + SG T ALVG+SGSGKSTVI+LL+RFYDPD
Sbjct: 158 IKGEIELRHISFKYPSRPDIQIFRDLNLTIHSGKTVALVGESGSGKSTVIALLQRFYDPD 337
Query: 381 AGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEG-ATDEEITTAITL 439
+GE+ +DG+ ++ LQL+W+R+Q+GLVSQEP+LF +IR NIAYGK G AT+ EI A L
Sbjct: 338 SGEITLDGIEIRQLQLKWLRQQMGLVSQEPVLFNDTIRANIAYGKGGIATEAEIIAAAEL 517
Query: 440 ANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESE 499
ANA +FI L QG DT+ G+ GTQLSGGQKQR+AIARAI+K+PKILLLDEATSALDAESE
Sbjct: 518 ANAHRFISGLQQGYDTIVGERGTQLSGGQKQRVAIARAIIKSPKILLLDEATSALDAESE 697
Query: 500 RIVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQL 559
R+VQ+AL+K+++ RTTVVVAHRL+TI+NAD+IAVV+ G IVE+G H L DG Y+ L
Sbjct: 698 RVVQDALDKVMVNRTTVVVAHRLSTIKNADVIAVVKNGVIVEKGRHETLINVKDGFYASL 877
Query: 560 IRL 562
++L
Sbjct: 878 VQL 886
>TC90210 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japonica},
partial (26%)
Length = 1158
Score = 390 bits (1003), Expect(2) = e-113
Identities = 210/391 (53%), Positives = 285/391 (72%), Gaps = 4/391 (1%)
Frame = +2
Query: 450 PQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKI 509
PQGLDTM G +GTQLSGGQKQRIAIARAILKNP+ILLLDEATSALDAESER+VQEAL++I
Sbjct: 2 PQGLDTMVGDHGTQLSGGQKQRIAIARAILKNPRILLLDEATSALDAESERVVQEALDRI 181
Query: 510 ILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEA 569
++ RTTVVVAHRL+T+RNAD+IAV+ +GK+VE+GTHS L DP+GAYSQLIRLQE + E+
Sbjct: 182 MVNRTTVVVAHRLSTVRNADMIAVIHRGKMVEKGTHSELLKDPEGAYSQLIRLQEVNKES 361
Query: 570 EGSRKSEADKLGDNLNIDSHMAGSSTQRTSFVRSISQTSSV--SHRHSQSLR-GLSGEIV 626
E + + + + S+QR S RSIS+ SS+ S RHS S+ GL +
Sbjct: 362 EETTDHHGKR-----ELSAESFRQSSQRKSLQRSISRGSSIGNSSRHSFSVSFGLPTGVN 526
Query: 627 ESDIEQGQLDNK-KKPKVSIWRLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVIS 685
+D + ++ K K+ +V + RLA LNKPEIPV+L+G++AAI NGV+ PIFG L S+VI
Sbjct: 527 VADPDLEKVPTKEKEQEVPLRRLASLNKPEIPVLLIGSLAAIANGVILPIFGVLISSVIK 706
Query: 686 MFYKPPEQQRKESRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQ 745
FY+P ++ +K+S+FW+++F+ LGL +LV++P + +FF +AG KLI+RIR L FEK+V+
Sbjct: 707 TFYEPFDEMKKDSKFWAIMFMLLGLASLVVIPARGYFFSVAGCKLIQRIRLLCFEKVVNM 886
Query: 746 EISWFDDPSHSRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDT 805
E+ W+D+P +S SGAVGARLS DA++V++LVGD
Sbjct: 887 EVGWYDEPENS---------------------------SGAVGARLSADAASVRALVGDA 985
Query: 806 MALIVQNISTVIAGLVIAFTANWILAFIVLV 836
+ L+VQN+++ +AGL+IAF A+W LA I+LV
Sbjct: 986 LGLLVQNLASALAGLIIAFIASWQLALIILV 1078
Score = 162 bits (411), Expect = 6e-40
Identities = 81/113 (71%), Positives = 94/113 (82%)
Frame = +2
Query: 1134 PDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRV 1193
P G DT VG+ GTQLSGGQKQRIAIAR +LKNP+ILLLDEATSALDAESER+VQEALDR+
Sbjct: 2 PQGLDTMVGDHGTQLSGGQKQRIAIARAILKNPRILLLDEATSALDAESERVVQEALDRI 181
Query: 1194 SVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELMRITDGVYASLVAL 1246
VNRTTVVVAHRL+T+R AD IAVI G + EKG H EL++ +G Y+ L+ L
Sbjct: 182 MVNRTTVVVAHRLSTVRNADMIAVIHRGKMVEKGTHSELLKDPEGAYSQLIRL 340
Score = 82.0 bits (201), Expect = 1e-15
Identities = 46/174 (26%), Positives = 98/174 (55%), Gaps = 2/174 (1%)
Frame = +2
Query: 27 KQKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFGSSNPADAIKQ 86
+Q+VP L + + ++ +++IG+++A+ANG+ P+ + + +VI F P D +K+
Sbjct: 569 EQEVPLRRLASL-NKPEIPVLLIGSLAAIANGVILPIFGVLISSVIKTF--YEPFDEMKK 739
Query: 87 VSKV-SLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTETNT 145
SK +++F+ L + S + + + V G + RIR L + ++ ++ ++D N+
Sbjct: 740 DSKFWAIMFMLLGLASLVVIPARGYFFSVAGCKLIQRIRLLCFEKVVNMEVGWYDEPENS 919
Query: 146 GEVIG-RMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLAC 198
+G R+S D ++ +G+ +G Q ++ G ++AFI W+LA+++L C
Sbjct: 920 SGAVGARLSADAASVRALVGDALGLLVQNLASALAGLIIAFIASWQLALIILVC 1081
Score = 38.9 bits (89), Expect(2) = e-113
Identities = 17/30 (56%), Positives = 23/30 (76%)
Frame = +3
Query: 833 IVLVLTPMILMQGIVQMKFLKGFSADAKVM 862
+ L + P+I + G VQMKF+KGFS DAK+M
Sbjct: 1068 LFLCVIPLIGLNGYVQMKFMKGFSGDAKMM 1157
>BG587938 homologue to GP|22655312|g putative ABC transporter {Arabidopsis
thaliana}, partial (19%)
Length = 780
Score = 313 bits (803), Expect = 2e-85
Identities = 160/246 (65%), Positives = 198/246 (80%)
Frame = +3
Query: 973 KAKDSAASIFEIIDSKPDIDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLS 1032
KA+ +AA IF IID +P ID +S +G+ ETV G +EL++V+F+YP+RP++ I D SLS
Sbjct: 39 KARVAAAKIFRIIDHQPGIDRNSESGLELETVTGLVELKNVDFSYPSRPEVLILNDFSLS 218
Query: 1033 IPSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQE 1092
+P+ KTIALVG SGSGKSTV+SL+ERFYDP SG+++LDG D+KT +L WLRQQ+GLV QE
Sbjct: 219 VPAGKTIALVGSSGSGKSTVVSLIERFYDPTSGQVMLDGHDIKTLKLKWLRQQIGLVSQE 398
Query: 1093 PILFNESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQ 1152
P LF +IR NI G+ A + EI AA ANAHSFI LP+G++T VGERG QLSGGQ
Sbjct: 399 PALFATTIRENILLGRP-DANQVEIEEAARVANAHSFIIKLPEGFETQVGERGLQLSGGQ 575
Query: 1153 KQRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGA 1212
KQRIAIAR MLKNP ILLLDEATSALD+ESE++VQEALDR + RTT+V+AHRL+TIR A
Sbjct: 576 KQRIAIARAMLKNPAILLLDEATSALDSESEKLVQEALDRFMIGRTTLVIAHRLSTIRKA 755
Query: 1213 DTIAVI 1218
D +AVI
Sbjct: 756 DLVAVI 773
Score = 276 bits (706), Expect = 4e-74
Identities = 135/216 (62%), Positives = 176/216 (80%)
Frame = +3
Query: 321 IKGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPD 380
+ G +EL++V F YP+RP+V I + FSL VP+G T ALVG SGSGKSTV+SL+ERFYDP
Sbjct: 132 VTGLVELKNVDFSYPSRPEVLILNDFSLSVPAGKTIALVGSSGSGKSTVVSLIERFYDPT 311
Query: 381 AGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLA 440
+G+V++DG ++K L+L+W+R+QIGLVSQEP LF T+IRENI G+ A EI A +A
Sbjct: 312 SGQVMLDGHDIKTLKLKWLRQQIGLVSQEPALFATTIRENILLGRPDANQVEIEEAARVA 491
Query: 441 NAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESER 500
NA FI KLP+G +T G+ G QLSGGQKQRIAIARA+LKNP ILLLDEATSALD+ESE+
Sbjct: 492 NAHSFIIKLPEGFETQVGERGLQLSGGQKQRIAIARAMLKNPAILLLDEATSALDSESEK 671
Query: 501 IVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQ 536
+VQEAL++ ++ RTT+V+AHRL+TIR AD++AV+ +
Sbjct: 672 LVQEALDRFMIGRTTLVIAHRLSTIRKADLVAVIHK 779
>BG646029 similar to GP|14715462|d CjMDR1 {Coptis japonica}, partial (16%)
Length = 680
Score = 261 bits (666), Expect(2) = 1e-74
Identities = 129/189 (68%), Positives = 159/189 (83%)
Frame = -3
Query: 1053 ISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGA 1112
ISLL+RFYDP+SG+I LDG +++ +L W RQQMGLV QEP+LFN++IRANI YGK G A
Sbjct: 606 ISLLQRFYDPDSGQIKLDGTEIQKLQLKWFRQQMGLVSQEPVLFNDTIRANIAYGKGGNA 427
Query: 1113 TEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLD 1172
TE E+IAAA ANAH+FIS+L GYDT VGERG QLSGGQKQ +AIAR ++ P+ILLLD
Sbjct: 426 TEAEVIAAAELANAHNFISSLQQGYDTIVGERGIQLSGGQKQPVAIARAIVNRPRILLLD 247
Query: 1173 EATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDEL 1232
EATSALDAESE++VQ+ALDRV V+RTT+VVAHRL+TI+GA++IAV+KNG + EKG+HD L
Sbjct: 246 EATSALDAESEKVVQDALDRVRVDRTTIVVAHRLSTIKGANSIAVVKNGVIEEKGKHDIL 67
Query: 1233 MRITDGVYA 1241
+ G YA
Sbjct: 66 IN-KGGTYA 43
Score = 223 bits (569), Expect(2) = 3e-62
Identities = 112/189 (59%), Positives = 148/189 (78%), Gaps = 1/189 (0%)
Frame = -3
Query: 370 ISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEG-A 428
ISLL+RFYDPD+G++ +DG ++ LQL+W R+Q+GLVSQEP+LF +IR NIAYGK G A
Sbjct: 606 ISLLQRFYDPDSGQIKLDGTEIQKLQLKWFRQQMGLVSQEPVLFNDTIRANIAYGKGGNA 427
Query: 429 TDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLD 488
T+ E+ A LANA FI L QG DT+ G+ G QLSGGQKQ +AIARAI+ P+ILLLD
Sbjct: 426 TEAEVIAAAELANAHNFISSLQQGYDTIVGERGIQLSGGQKQPVAIARAIVNRPRILLLD 247
Query: 489 EATSALDAESERIVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGL 548
EATSALDAESE++VQ+AL+++ + RTT+VVAHRL+TI+ A+ IAVV+ G I E+G H +
Sbjct: 246 EATSALDAESEKVVQDALDRVRVDRTTIVVAHRLSTIKGANSIAVVKNGVIEEKGKHD-I 70
Query: 549 TMDPDGAYS 557
++ G Y+
Sbjct: 69 LINKGGTYA 43
Score = 38.9 bits (89), Expect(2) = 1e-74
Identities = 19/24 (79%), Positives = 22/24 (91%)
Frame = -2
Query: 1029 LSLSIPSAKTIALVGESGSGKSTV 1052
LSL+I S +T+ALVGESGSGKSTV
Sbjct: 679 LSLTIHSGQTVALVGESGSGKSTV 608
Score = 35.0 bits (79), Expect(2) = 3e-62
Identities = 17/23 (73%), Positives = 19/23 (81%)
Frame = -2
Query: 347 SLFVPSGTTTALVGQSGSGKSTV 369
SL + SG T ALVG+SGSGKSTV
Sbjct: 676 SLTIHSGQTVALVGESGSGKSTV 608
>TC81811 similar to GP|6671365|gb|AAF23176.1| P-glycoprotein {Gossypium
hirsutum}, partial (17%)
Length = 850
Score = 276 bits (705), Expect = 5e-74
Identities = 139/218 (63%), Positives = 173/218 (78%)
Frame = +1
Query: 350 VPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQIGLVSQE 409
VPSG + ALVGQSGSGKS+VISL+ RFYDP +G+VLIDG ++ + L+ + + IGLV QE
Sbjct: 1 VPSGKSVALVGQSGSGKSSVISLILRFYDPTSGKVLIDGKDITRINLKSLTKHIGLVQQE 180
Query: 410 PILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQLSGGQK 469
P LF TSI ENI YGKEGA+D E+ A LANA FI LP+G T G+ G QLSGGQ+
Sbjct: 181 PALFATSIYENILYGKEGASDSEVIEAAKLANAHNFISALPEGYSTKVGERGVQLSGGQR 360
Query: 470 QRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLTTIRNAD 529
QR+AIARA+LKNP+ILLLDEATSALD ESERIVQ+AL++++ RTTV+VAHRL+TIRNAD
Sbjct: 361 QRVAIARAVLKNPEILLLDEATSALDVESERIVQQALDRLMQNRTTVMVAHRLSTIRNAD 540
Query: 530 IIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDN 567
I+V+Q GKI+E+GTHS L + DG Y +L+ LQ+ N
Sbjct: 541 QISVLQDGKIIEQGTHSSLIENKDGPYYKLVNLQQQQN 654
Score = 263 bits (673), Expect = 2e-70
Identities = 136/214 (63%), Positives = 168/214 (77%)
Frame = +1
Query: 1033 IPSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQE 1092
+PS K++ALVG+SGSGKS+VISL+ RFYDP SG++L+DG D+ L L + +GLV QE
Sbjct: 1 VPSGKSVALVGQSGSGKSSVISLILRFYDPTSGKVLIDGKDITRINLKSLTKHIGLVQQE 180
Query: 1093 PILFNESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQ 1152
P LF SI NI YGKEG A++ E+I AA ANAH+FIS LP+GY T VGERG QLSGGQ
Sbjct: 181 PALFATSIYENILYGKEG-ASDSEVIEAAKLANAHNFISALPEGYSTKVGERGVQLSGGQ 357
Query: 1153 KQRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGA 1212
+QR+AIAR +LKNP+ILLLDEATSALD ESERIVQ+ALDR+ NRTTV+VAHRL+TIR A
Sbjct: 358 RQRVAIARAVLKNPEILLLDEATSALDVESERIVQQALDRLMQNRTTVMVAHRLSTIRNA 537
Query: 1213 DTIAVIKNGAVAEKGRHDELMRITDGVYASLVAL 1246
D I+V+++G + E+G H L+ DG Y LV L
Sbjct: 538 DQISVLQDGKIIEQGTHSSLIENKDGPYYKLVNL 639
>TC78215 similar to GP|10177549|dbj|BAB10828. ABC transporter-like protein
{Arabidopsis thaliana}, partial (36%)
Length = 1176
Score = 232 bits (592), Expect(2) = 1e-60
Identities = 118/224 (52%), Positives = 158/224 (69%)
Frame = +1
Query: 1021 PDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLS 1080
P + K +++ + +ALVG SG GK+T+ +L+ERFYDP G IL++GV L
Sbjct: 34 P*YMVLKGINIKLQPGSKVALVGPSGGGKTTIANLIERFYDPTKGNILVNGVPLVEISHK 213
Query: 1081 WLRQQMGLVGQEPILFNESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTS 1140
L +++ +V QEP LFN SI NI YG +G + +I AA ANAH FISN P+ Y T
Sbjct: 214 HLHRKISIVSQEPTLFNCSIEENIAYGFDGKIEDADIENAAKMANAHEFISNFPEKYKTF 393
Query: 1141 VGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRTTV 1200
VGERG +LSGGQKQRIAIAR +L +PKILLLDEATSALDAESE +VQ+A+D + RT +
Sbjct: 394 VGERGIRLSGGQKQRIAIARALLMDPKILLLDEATSALDAESEYLVQDAMDSIMKGRTVL 573
Query: 1201 VVAHRLTTIRGADTIAVIKNGAVAEKGRHDELMRITDGVYASLV 1244
V+AHRL+T++ A+T+AV+ +G + E G HDEL+ +GVY +LV
Sbjct: 574 VIAHRLSTVKTANTVAVVSDGQIVESGTHDELLE-KNGVYTALV 702
Score = 221 bits (564), Expect(2) = 6e-58
Identities = 114/227 (50%), Positives = 159/227 (69%), Gaps = 1/227 (0%)
Frame = +1
Query: 338 PDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLR 397
P + G ++ + G+ ALVG SG GK+T+ +L+ERFYDP G +L++GV L + +
Sbjct: 34 P*YMVLKGINIKLQPGSKVALVGPSGGGKTTIANLIERFYDPTKGNILVNGVPLVEISHK 213
Query: 398 WIREQIGLVSQEPILFTTSIRENIAYGKEGAT-DEEITTAITLANAKKFIDKLPQGLDTM 456
+ +I +VSQEP LF SI ENIAYG +G D +I A +ANA +FI P+ T
Sbjct: 214 HLHRKISIVSQEPTLFNCSIEENIAYGFDGKIEDADIENAAKMANAHEFISNFPEKYKTF 393
Query: 457 AGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTV 516
G+ G +LSGGQKQRIAIARA+L +PKILLLDEATSALDAESE +VQ+A++ I+ RT +
Sbjct: 394 VGERGIRLSGGQKQRIAIARALLMDPKILLLDEATSALDAESEYLVQDAMDSIMKGRTVL 573
Query: 517 VVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQ 563
V+AHRL+T++ A+ +AVV G+IVE GTH L ++ +G Y+ L++ Q
Sbjct: 574 VIAHRLSTVKTANTVAVVSDGQIVESGTHDEL-LEKNGVYTALVKRQ 711
Score = 22.7 bits (47), Expect(2) = 6e-58
Identities = 8/14 (57%), Positives = 11/14 (78%)
Frame = +2
Query: 327 LRDVSFRYPARPDV 340
L DV F YP+RP++
Sbjct: 2 LNDVWFSYPSRPNI 43
Score = 20.8 bits (42), Expect(2) = 1e-60
Identities = 8/14 (57%), Positives = 11/14 (78%)
Frame = +2
Query: 1010 LQHVNFNYPTRPDI 1023
L V F+YP+RP+I
Sbjct: 2 LNDVWFSYPSRPNI 43
>TC86470 similar to PIR|F84919|F84919 glutathione-conjugate transporter AtMRP4
[imported] - Arabidopsis thaliana, partial (57%)
Length = 3175
Score = 222 bits (565), Expect = 8e-58
Identities = 207/942 (21%), Positives = 423/942 (43%), Gaps = 22/942 (2%)
Frame = +1
Query: 325 IELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEV 384
++++D +F + Q +L V G TA+VG GSGKS++++ + ++G+V
Sbjct: 154 VDVQDGTFSWDDEGLEQDLKNINLKVNKGELTAIVGTVGSGKSSLLASILGEMHRNSGKV 333
Query: 385 LIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKK 444
+ G Q WI+ +I ENI +G ++ I + +K
Sbjct: 334 QVCGSTAYVAQTSWIQNG-------------TIEENILFGLP-MNRQKYNEIIRVCCLEK 471
Query: 445 FIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAES-ERIVQ 503
++ + G T G+ G LSGGQKQRI +ARA+ ++ I LLD+ SA+DA + I +
Sbjct: 472 DLEMMEYGDQTEIGERGINLSGGQKQRIQLARAVYQDCDIYLLDDVFSAVDAHTGTEIFK 651
Query: 504 EALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQ 563
E + + +T V+V H++ + N D I V++ G IV+ G ++ L +D + L+
Sbjct: 652 ECVRGALKGKTIVLVTHQVDFLHNVDRIVVMRDGMIVQSGRYNDL-LDSGLDFGVLVAAH 828
Query: 564 EGDNEAEGSRKSEADKLGDNLNIDSHMAGSSTQRTSFVRSISQTSSVSHRHSQSLRGLSG 623
E E + + + L I + ++ + S+ Q +S +G S
Sbjct: 829 ETSMELVEQGAAVPGENSNKLMISKSASINNRETNGESNSLDQPNSA--------KGSSK 984
Query: 624 EIVESDIEQGQLDNKKKPKVSIWRLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAV 683
+ E + E G++ + G +A + V++ S +
Sbjct: 985 LVKEEERETGKVSFNIYKRYCTEAFG----------WAGILAVLFLSVLWQA-----SMM 1119
Query: 684 ISMFYKPPEQQRKESRFWS-----LLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLT 738
S ++ E + + ++ ++ + +V+++++ ++++ I G K + +
Sbjct: 1120 ASDYWLAFETSVERAEVFNPVVFISIYAAITIVSVILIVVRSYSVTIFGLKTAQIFFNQI 1299
Query: 739 FEKIVHQEISWFDDPSHSRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTV 798
I+H +S++D ++ SG + +R S D + V
Sbjct: 1300 LTSILHAPMSFYD-----------------------------TTPSGRILSRASTDQTNV 1392
Query: 799 KSLVGDTMALIVQNISTVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSAD 858
+ + +V TVI+ ++I +W AF+++ L + + + + +
Sbjct: 1393 DIFIPLFINFVVAMYITVISIVIITCQNSWPTAFLLIPLVWLNIWYRGYFLSTSRELTRL 1572
Query: 859 AKVMYEEASQVANDAVSSIRTVASFCAESKV-------------MDMYSKKCLGPAKQGV 905
+ ++++S + TV +F + + MD ++ A G
Sbjct: 1573 DSITKAPVIVHFSESISGVMTVRAFRKQKEFRLENFKRVNSNLRMDFHNYS--SNAWLGF 1746
Query: 906 RLGLVSGIGFGCS--FLVLYCTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQ 963
RL L+ + F S F++L +N + + +G + + +F +A+ M+ ++
Sbjct: 1747 RLELLGSLVFCLSALFMILLPSNIIKPENVGLSLSYGLSLNSVLF---WAIYMSCFIENK 1917
Query: 964 TTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTRETVVGDIELQHVNFNYPTRPDI 1023
++ + + + + I D P + G ++++ + Y RP+
Sbjct: 1918 MVSVERIKQFSNIPSEAAWNIKDRSPPPNWPGQ---------GHVDIKDLQVRY--RPNT 2064
Query: 1024 Q-IFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWL 1082
+ K ++LSI + + +VG +GSGKST+I + R +P G+I++DG+D+ L L
Sbjct: 2065 PLVLKGITLSISGGEKVGVVGRTGSGKSTLIQVFFRLVEPTGGKIIIDGIDICALGLHDL 2244
Query: 1083 RQQMGLVGQEPILFNESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVG 1142
R + G++ QEP+LF ++R+NI G T+DEI + + +++ P+ D+ V
Sbjct: 2245 RSRFGIIPQEPVLFEGTVRSNID--PTGQYTDDEIWKSLDRCQLKDTVASKPEKLDSLVV 2418
Query: 1143 ERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRTTVVV 1202
+ G S GQ+Q + + R MLK ++L +DEAT+++D++++ ++Q+ + RT + +
Sbjct: 2419 DNGDNWSVGQRQLLCLGRVMLKQSRLLFMDEATASVDSQTDAVIQKIIREDFAARTIISI 2598
Query: 1203 AHRLTTIRGADTIAVIKNGAVAEKGRHDELMRITDGVYASLV 1244
AHR+ T+ D + V+ G E + L++ ++A+LV
Sbjct: 2599 AHRIPTVMDCDRVLVVDAGRAKEFDKPSNLLQ-RQSLFAALV 2721
Score = 147 bits (371), Expect = 3e-35
Identities = 131/540 (24%), Positives = 244/540 (44%), Gaps = 58/540 (10%)
Frame = +1
Query: 90 VSLLFVYLAIGSGIASFLQVTCWMVT--GERQAARIRSLYLKTILQQDIAFFDTETNTGE 147
V + +Y AI + V + VT G + A + L +IL ++F+DT T +G
Sbjct: 1180 VVFISIYAAITIVSVILIVVRSYSVTIFGLKTAQIFFNQILTSILHAPMSFYDT-TPSGR 1356
Query: 148 VIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIK--------GWRLAIVLLACV 199
++ R S D V F L NF + I W A +L+ V
Sbjct: 1357 ILSRASTDQT--------NVDIFIPLFINFVVAMYITVISIVIITCQNSWPTAFLLIPLV 1512
Query: 200 PC-VAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKA----IEKY 254
+ G F+S +++ I A +++ + TV +F +K+ ++
Sbjct: 1513 WLNIWYRGYFLS-TSRELTRLDSITKAPVIVHFSESISGVMTVRAFRKQKEFRLENFKRV 1689
Query: 255 NSKIKIAYTTMVKQGIVSGFGIGMLTFIAFC-----------------TYGLAMWYGSKL 297
NS +++ + + GF + +L + FC GL++ YG L
Sbjct: 1690 NSNLRMDFHNYSSNAWL-GFRLELLGSLVFCLSALFMILLPSNIIKPENVGLSLSYGLSL 1866
Query: 298 -------VIEKGYNGGTVMTVIIALMTGGII------------------KGDIELRDVSF 332
+ + +++V I +G ++++D+
Sbjct: 1867 NSVLFWAIYMSCFIENKMVSVERIKQFSNIPSEAAWNIKDRSPPPNWPGQGHVDIKDLQV 2046
Query: 333 RYPARPDVQ-IFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNL 391
RY RP+ + G +L + G +VG++GSGKST+I + R +P G+++IDG+++
Sbjct: 2047 RY--RPNTPLVLKGITLSISGGEKVGVVGRTGSGKSTLIQVFFRLVEPTGGKIIIDGIDI 2220
Query: 392 KNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQ 451
L L +R + G++ QEP+LF ++R NI + TD+EI ++ K + P+
Sbjct: 2221 CALGLHDLRSRFGIIPQEPVLFEGTVRSNIDPTGQ-YTDDEIWKSLDRCQLKDTVASKPE 2397
Query: 452 GLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIIL 511
LD++ NG S GQ+Q + + R +LK ++L +DEAT+++D++++ ++Q+ + +
Sbjct: 2398 KLDSLVVDNGDNWSVGQRQLLCLGRVMLKQSRLLFMDEATASVDSQTDAVIQKIIREDFA 2577
Query: 512 KRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEAEG 571
RT + +AHR+ T+ + D + VV G+ E S L + ++ L+ QE N + G
Sbjct: 2578 ARTIISIAHRIPTVMDCDRVLVVDAGRAKEFDKPSNL-LQRQSLFAALV--QEYANRSTG 2748
Score = 97.4 bits (241), Expect = 3e-20
Identities = 70/246 (28%), Positives = 126/246 (50%), Gaps = 3/246 (1%)
Frame = +1
Query: 1008 IELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGRI 1067
+++Q F++ Q K+++L + + A+VG GSGKS++++ + NSG++
Sbjct: 154 VDVQDGTFSWDDEGLEQDLKNINLKVNKGELTAIVGTVGSGKSSLLASILGEMHRNSGKV 333
Query: 1068 LLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGATE-DEIIAAANAANA 1126
+ G + SW++ N +I NI +G + +EII
Sbjct: 334 QVCGSTAYVAQTSWIQ-------------NGTIEENILFGLPMNRQKYNEII---RVCCL 465
Query: 1127 HSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAES-ERI 1185
+ + G T +GERG LSGGQKQRI +AR + ++ I LLD+ SA+DA + I
Sbjct: 466 EKDLEMMEYGDQTEIGERGINLSGGQKQRIQLARAVYQDCDIYLLDDVFSAVDAHTGTEI 645
Query: 1186 VQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELMRITDGV-YASLV 1244
+E + +T V+V H++ + D I V+++G + + GR+++L + G+ + LV
Sbjct: 646 FKECVRGALKGKTIVLVTHQVDFLHNVDRIVVMRDGMIVQSGRYNDL--LDSGLDFGVLV 819
Query: 1245 ALHSSA 1250
A H ++
Sbjct: 820 AAHETS 837
>TC77935 similar to PIR|E85023|E85023 probable P-glycoprotein-like protein
[imported] - Arabidopsis thaliana, partial (12%)
Length = 869
Score = 220 bits (561), Expect = 2e-57
Identities = 111/155 (71%), Positives = 132/155 (84%)
Frame = +2
Query: 1095 LFNESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQ 1154
LFN+++RANI YGK G ATE EI+AAA ANAH FI +L GYDT VGERG QLSGGQKQ
Sbjct: 65 LFNDTVRANIAYGKGGDATEAEIVAAAELANAHQFIGSLQKGYDTIVGERGIQLSGGQKQ 244
Query: 1155 RIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADT 1214
R+AIAR ++KNPKILLLDEATSALDAESE++VQ+ALDRV V RTT++VAHRL+TI+GAD
Sbjct: 245 RVAIARAIVKNPKILLLDEATSALDAESEKVVQDALDRVMVERTTIIVAHRLSTIKGADL 424
Query: 1215 IAVIKNGAVAEKGRHDELMRITDGVYASLVALHSS 1249
IAV+KNG +AEKG+H+ L+ G YASLVALH+S
Sbjct: 425 IAVVKNGVIAEKGKHEALLH-KGGDYASLVALHTS 526
Score = 181 bits (460), Expect = 1e-45
Identities = 93/157 (59%), Positives = 122/157 (77%), Gaps = 1/157 (0%)
Frame = +2
Query: 412 LFTTSIRENIAYGKEG-ATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQ 470
LF ++R NIAYGK G AT+ EI A LANA +FI L +G DT+ G+ G QLSGGQKQ
Sbjct: 65 LFNDTVRANIAYGKGGDATEAEIVAAAELANAHQFIGSLQKGYDTIVGERGIQLSGGQKQ 244
Query: 471 RIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLTTIRNADI 530
R+AIARAI+KNPKILLLDEATSALDAESE++VQ+AL++++++RTT++VAHRL+TI+ AD+
Sbjct: 245 RVAIARAIVKNPKILLLDEATSALDAESEKVVQDALDRVMVERTTIIVAHRLSTIKGADL 424
Query: 531 IAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDN 567
IAVV+ G I E+G H L + G Y+ L+ L D+
Sbjct: 425 IAVVKNGVIAEKGKHEAL-LHKGGDYASLVALHTSDS 532
>TC89340 weakly similar to GP|17427488|emb|CAD14006. PROBABLE COMPOSITE
ATP-BINDING TRANSMEMBRANE ABC TRANSPORTER PROTEIN
{Ralstonia solanacearum}, partial (32%)
Length = 795
Score = 205 bits (521), Expect = 1e-52
Identities = 116/264 (43%), Positives = 162/264 (60%)
Frame = +1
Query: 977 SAASIFEIIDSKPDIDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSA 1036
S + E+ KP + S A R+ G+I ++V FNY P + KDL
Sbjct: 16 SGERLLELFKVKPTVVDSPTAEPLRDCN-GNIRWKNVGFNYD--PKNKALKDLDFECKPG 186
Query: 1037 KTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILF 1096
T A VGESG GKSTV L+ R+Y+ SG I +DG D+K + +R +G+V Q+ LF
Sbjct: 187 STTAFVGESGGGKSTVFRLMFRYYNTTSGSIEIDGRDVKDLTIDSVRSYIGVVPQDTTLF 366
Query: 1097 NESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRI 1156
NES+ N+ Y + AT++EI A AA+ H I + PD YDT VGERG +LSGG++QR+
Sbjct: 367 NESLMYNLRYARPE-ATDEEIYDACRAASIHDRIMSFPDRYDTKVGERGLKLSGGERQRV 543
Query: 1157 AIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIA 1216
AIART++K+PKI++LDEATSALDA +E+ +Q+ L + RT +++AHRL+TI AD I
Sbjct: 544 AIARTIIKSPKIIMLDEATSALDAHTEQQIQDRLGHLGQGRTLLIIAHRLSTITHADQII 723
Query: 1217 VIKNGAVAEKGRHDELMRITDGVY 1240
V + KG H EL+ DG +
Sbjct: 724 VPQQWDY**KGHHKELVMPMDGTH 795
Score = 180 bits (456), Expect = 4e-45
Identities = 100/234 (42%), Positives = 143/234 (60%)
Frame = +1
Query: 323 GDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAG 382
G+I ++V F Y P + G+TTA VG+SG GKSTV L+ R+Y+ +G
Sbjct: 100 GNIRWKNVGFNYD--PKNKALKDLDFECKPGSTTAFVGESGGGKSTVFRLMFRYYNTTSG 273
Query: 383 EVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANA 442
+ IDG ++K+L + +R IG+V Q+ LF S+ N+ Y + ATDEEI A A+
Sbjct: 274 SIEIDGRDVKDLTIDSVRSYIGVVPQDTTLFNESLMYNLRYARPEATDEEIYDACRAASI 453
Query: 443 KKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIV 502
I P DT G+ G +LSGG++QR+AIAR I+K+PKI++LDEATSALDA +E+ +
Sbjct: 454 HDRIMSFPDRYDTKVGERGLKLSGGERQRVAIARTIIKSPKIIMLDEATSALDAHTEQQI 633
Query: 503 QEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAY 556
Q+ L + RT +++AHRL+TI +AD I V QQ +G H L M DG +
Sbjct: 634 QDRLGHLGQGRTLLIIAHRLSTITHADQIIVPQQWDY**KGHHKELVMPMDGTH 795
>TC83850 weakly similar to GP|7300071|gb|AAF55241.1| CG4225 gene product
{Drosophila melanogaster}, partial (23%)
Length = 816
Score = 202 bits (513), Expect = 9e-52
Identities = 105/211 (49%), Positives = 148/211 (69%)
Frame = +2
Query: 1034 PSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEP 1093
P KT A+VGESGSGKST + LL RFYD G + +DG D+K +L LR+ +G+V Q+
Sbjct: 53 PGTKT-AIVGESGSGKSTCLKLLFRFYDVQEGAVTVDGHDIKHLKLDSLRRNIGVVPQDT 229
Query: 1094 ILFNESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQK 1153
+LFN +I N+ Y A++ ++ A AAN H I N PDGY+T VGERG +LSGG++
Sbjct: 230 VLFNATIMYNLLYANPK-ASQADVFEACKAANIHERILNFPDGYETKVGERGLKLSGGER 406
Query: 1154 QRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGAD 1213
QRIAIAR +LK+ +ILLLDEAT++LD+ +ER +Q+AL+RV+ RTT+ +AHRL+TI +D
Sbjct: 407 QRIAIARAILKDARILLLDEATASLDSHTERQIQDALERVTAGRTTITIAHRLSTITTSD 586
Query: 1214 TIAVIKNGAVAEKGRHDELMRITDGVYASLV 1244
I V+ + E+G H EL+ + G Y ++V
Sbjct: 587 QIVVLHKSKIVERGTHSELLAL-GGRYHAMV 676
Score = 200 bits (508), Expect = 3e-51
Identities = 103/205 (50%), Positives = 142/205 (69%)
Frame = +2
Query: 344 DGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQI 403
DG S V GT TA+VG+SGSGKST + LL RFYD G V +DG ++K+L+L +R I
Sbjct: 29 DGVSFTVAPGTKTAIVGESGSGKSTCLKLLFRFYDVQEGAVTVDGHDIKHLKLDSLRRNI 208
Query: 404 GLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQ 463
G+V Q+ +LF +I N+ Y A+ ++ A AN + I P G +T G+ G +
Sbjct: 209 GVVPQDTVLFNATIMYNLLYANPKASQADVFEACKAANIHERILNFPDGYETKVGERGLK 388
Query: 464 LSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLT 523
LSGG++QRIAIARAILK+ +ILLLDEAT++LD+ +ER +Q+ALE++ RTT+ +AHRL+
Sbjct: 389 LSGGERQRIAIARAILKDARILLLDEATASLDSHTERQIQDALERVTAGRTTITIAHRLS 568
Query: 524 TIRNADIIAVVQQGKIVERGTHSGL 548
TI +D I V+ + KIVERGTHS L
Sbjct: 569 TITTSDQIVVLHKSKIVERGTHSEL 643
>AL382570 weakly similar to GP|10241621|em putative P-glycoprotein {Oryza
sativa}, partial (17%)
Length = 481
Score = 181 bits (458), Expect = 2e-45
Identities = 93/151 (61%), Positives = 115/151 (75%)
Frame = +3
Query: 1049 KSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGK 1108
KS+V++LL +FYDP G++L+DG DL+ + L WLR Q+GLV QEP+LFN SIR NI YG
Sbjct: 3 KSSVLALLLKFYDPVVGKVLIDGKDLREYNLRWLRTQIGLVQQEPLLFNCSIRENICYGN 182
Query: 1109 EGGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKI 1168
G A E EI+ A AN H F+SNLP+GY+T VGE+G QLSGGQKQRIAIART+LK P I
Sbjct: 183 NG-AFESEIVEVAREANIHEFVSNLPNGYNTVVGEKGCQLSGGQKQRIAIARTLLKKPAI 359
Query: 1169 LLLDEATSALDAESERIVQEALDRVSVNRTT 1199
LLLDEATSALDA SER + A+ +++ T
Sbjct: 360 LLLDEATSALDA*SERTIVNAIKAMNLKEET 452
Score = 172 bits (435), Expect = 1e-42
Identities = 88/150 (58%), Positives = 108/150 (71%)
Frame = +3
Query: 366 KSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGK 425
KS+V++LL +FYDP G+VLIDG +L+ LRW+R QIGLV QEP+LF SIRENI YG
Sbjct: 3 KSSVLALLLKFYDPVVGKVLIDGKDLREYNLRWLRTQIGLVQQEPLLFNCSIRENICYGN 182
Query: 426 EGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKIL 485
GA + EI AN +F+ LP G +T+ G+ G QLSGGQKQRIAIAR +LK P IL
Sbjct: 183 NGAFESEIVEVAREANIHEFVSNLPNGYNTVVGEKGCQLSGGQKQRIAIARTLLKKPAIL 362
Query: 486 LLDEATSALDAESERIVQEALEKIILKRTT 515
LLDEATSALDA SER + A++ + LK T
Sbjct: 363 LLDEATSALDA*SERTIVNAIKAMNLKEET 452
>AW208110 similar to GP|14715462|d CjMDR1 {Coptis japonica}, partial (11%)
Length = 449
Score = 175 bits (444), Expect = 9e-44
Identities = 88/146 (60%), Positives = 109/146 (74%)
Frame = +2
Query: 839 PMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIRTVASFCAESKVMDMYSKKCL 898
P++ + G VQ+KFLKGFSADAK +YEEASQVANDAV IRTV+SFCAE KVM++Y +KC
Sbjct: 11 PLLGLDGYVQVKFLKGFSADAKKLYEEASQVANDAVGCIRTVSSFCAEEKVMELYEQKCE 190
Query: 899 GPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMTA 958
GP K+G+R G++SG+GFG S +LY A FY G+ LV+ GK+TF++VF V FAL M A
Sbjct: 191 GPIKKGIRRGIISGLGFGLSCFLLYAVYACCFYAGARLVEDGKSTFSDVFLVIFALGMAA 370
Query: 959 IAVSQTTTLAPDTNKAKDSAASIFEI 984
VSQ TL PD AK + ASIF I
Sbjct: 371 SGVSQLGTLVPDLINAKSATASIFAI 448
Score = 65.1 bits (157), Expect = 2e-10
Identities = 36/116 (31%), Positives = 57/116 (49%)
Frame = +2
Query: 200 PCVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIK 259
P + + G + S+ + Y EA V + VG IRTV+SF E+K +E Y K +
Sbjct: 11 PLLGLDGYVQVKFLKGFSADAKKLYEEASQVANDAVGCIRTVSSFCAEEKVMELYEQKCE 190
Query: 260 IAYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIAL 315
+++GI+SG G G+ F+ + Y + G++LV + V VI AL
Sbjct: 191 GPIKKGIRRGIISGLGFGLSCFLLYAVYACCFYAGARLVEDGKSTFSDVFLVIFAL 358
>AL383696 homologue to PIR|T47671|T4 P-glycoprotein-like - Arabidopsis
thaliana, partial (9%)
Length = 391
Score = 147 bits (370), Expect = 3e-35
Identities = 73/130 (56%), Positives = 95/130 (72%)
Frame = +1
Query: 1025 IFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQ 1084
+ + SL + +T+A+VG SGSGKST+ISL+ERFYDP +G++LLDG DLK + L WLR
Sbjct: 4 VLSNFSLKVNGGQTVAVVGVSGSGKSTIISLIERFYDPVAGQVLLDGRDLKLYNLRWLRS 183
Query: 1085 QMGLVGQEPILFNESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVGER 1144
+GL+ QEPI+F+ +IR NI Y + A+E E+ AA ANAH FIS+LP GYDT VG R
Sbjct: 184 HLGLIQQEPIIFSTTIRENIIYARH-NASEAEMKEAARIANAHHFISSLPHGYDTHVGMR 360
Query: 1145 GTQLSGGQKQ 1154
G L+ GQKQ
Sbjct: 361 GVDLTPGQKQ 390
Score = 145 bits (367), Expect = 7e-35
Identities = 71/129 (55%), Positives = 90/129 (69%)
Frame = +1
Query: 342 IFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIRE 401
+ FSL V G T A+VG SGSGKST+ISL+ERFYDP AG+VL+DG +LK LRW+R
Sbjct: 4 VLSNFSLKVNGGQTVAVVGVSGSGKSTIISLIERFYDPVAGQVLLDGRDLKLYNLRWLRS 183
Query: 402 QIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNG 461
+GL+ QEPI+F+T+IRENI Y + A++ E+ A +ANA FI LP G DT G G
Sbjct: 184 HLGLIQQEPIIFSTTIRENIIYARHNASEAEMKEAARIANAHHFISSLPHGYDTHVGMRG 363
Query: 462 TQLSGGQKQ 470
L+ GQKQ
Sbjct: 364 VDLTPGQKQ 390
>TC87490 similar to PIR|T52080|T52080 multi resistance protein [imported] -
Arabidopsis thaliana, partial (19%)
Length = 927
Score = 145 bits (366), Expect = 1e-34
Identities = 81/240 (33%), Positives = 138/240 (56%)
Frame = +1
Query: 321 IKGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPD 380
+ G I+L D+ RY + + G S P G +VG++GSGKST+I L R +P
Sbjct: 154 VNGTIQLIDLKVRYKENLPM-VLHGVSCTFPGGKKIGIVGRTGSGKSTLIQALFRLVEPA 330
Query: 381 AGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLA 440
AG +LID +++ + L +R + ++ Q+P LF +IR N+ +E + D+EI A+ +
Sbjct: 331 AGSILIDNIDISGIGLHDLRSHLSIIPQDPTLFEGTIRGNLDPLEEHS-DKEIWEALDKS 507
Query: 441 NAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESER 500
+ I + Q LDT QNG S GQ+Q +A+ RA+LK KIL+LDEAT+++D+ ++
Sbjct: 508 QLGEIIREKGQKLDTPVLQNGDNWSVGQRQLVALGRALLKQSKILVLDEATASVDSATDN 687
Query: 501 IVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLI 560
++Q+ + + T +AHR+ T+ ++D++ V+ G + E T L D + +L+
Sbjct: 688 LIQKVIREEFRDCTVCTIAHRIPTVIDSDLVLVLNDGLLAEFDTPLRLLEDKSSMFLKLV 867
Score = 130 bits (326), Expect = 4e-30
Identities = 78/246 (31%), Positives = 132/246 (52%)
Frame = +1
Query: 1004 VVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPN 1063
V G I+L + Y + + +S + P K I +VG +GSGKST+I L R +P
Sbjct: 154 VNGTIQLIDLKVRYKENLPM-VLHGVSCTFPGGKKIGIVGRTGSGKSTLIQALFRLVEPA 330
Query: 1064 SGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGATEDEIIAAANA 1123
+G IL+D +D+ L LR + ++ Q+P LF +IR N+ +E ++ EI A +
Sbjct: 331 AGSILIDNIDISGIGLHDLRSHLSIIPQDPTLFEGTIRGNLDPLEEH--SDKEIWEALDK 504
Query: 1124 ANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESE 1183
+ I DT V + G S GQ+Q +A+ R +LK KIL+LDEAT+++D+ ++
Sbjct: 505 SQLGEIIREKGQKLDTPVLQNGDNWSVGQRQLVALGRALLKQSKILVLDEATASVDSATD 684
Query: 1184 RIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELMRITDGVYASL 1243
++Q+ + + T +AHR+ T+ +D + V+ +G +AE L+ ++ L
Sbjct: 685 NLIQKVIREEFRDCTVCTIAHRIPTVIDSDLVLVLNDGLLAEFDTPLRLLEDKSSMFLKL 864
Query: 1244 VALHSS 1249
V +SS
Sbjct: 865 VTEYSS 882
>TC92097 similar to GP|22553016|emb|CAD44995. multidrug-resistance related
protein {Arabidopsis thaliana}, partial (25%)
Length = 1011
Score = 145 bits (366), Expect = 1e-34
Identities = 82/222 (36%), Positives = 135/222 (59%), Gaps = 2/222 (0%)
Frame = +2
Query: 322 KGDIELRDVSFRYPARPDVQ-IFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPD 380
KG I+L+ + RY RP+ + G G+ +VG++GSGKST+IS L R +P
Sbjct: 347 KGKIDLQGLEIRY--RPNSPLVLKGIICTFKEGSRVGVVGRTGSGKSTLISALFRLVEPS 520
Query: 381 AGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEGA-TDEEITTAITL 439
G++LIDG+N+ ++ L+ +R ++ ++ QEP LF SIR N+ G +D+EI A+
Sbjct: 521 RGDILIDGINICSIGLKDLRTKLSIIPQEPTLFKGSIRTNL--DPLGLYSDDEIWKAVEK 694
Query: 440 ANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESE 499
K+ I KLP LD+ G S GQ+Q + R +LK +IL+LDEAT+++D+ ++
Sbjct: 695 CQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATD 874
Query: 500 RIVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVE 541
I+Q + + + T + VAHR+ T+ ++D++ V+ GK+VE
Sbjct: 875 AILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVE 1000
Score = 133 bits (335), Expect = 4e-31
Identities = 78/221 (35%), Positives = 124/221 (55%), Gaps = 1/221 (0%)
Frame = +2
Query: 1006 GDIELQHVNFNY-PTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNS 1064
G I+LQ + Y P P + K + + + +VG +GSGKST+IS L R +P+
Sbjct: 350 GKIDLQGLEIRYRPNSP--LVLKGIICTFKEGSRVGVVGRTGSGKSTLISALFRLVEPSR 523
Query: 1065 GRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGATEDEIIAAANAA 1124
G IL+DG+++ + L LR ++ ++ QEP LF SIR N+ G ++DEI A
Sbjct: 524 GDILIDGINICSIGLKDLRTKLSIIPQEPTLFKGSIRTNLD--PLGLYSDDEIWKAVEKC 697
Query: 1125 NAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESER 1184
IS LP D+SV + G S GQ+Q + R +LK +IL+LDEAT+++D+ ++
Sbjct: 698 QLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDA 877
Query: 1185 IVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAE 1225
I+Q + + T + VAHR+ T+ +D + V+ G + E
Sbjct: 878 ILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVE 1000
>BM813979 homologue to PIR|E85023|E85 probable P-glycoprotein-like protein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 434
Score = 144 bits (363), Expect = 2e-34
Identities = 70/97 (72%), Positives = 83/97 (85%)
Frame = +3
Query: 1037 KTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILF 1096
+T+ALVGESGSGKSTVI+LL+RFYDP+SG I LDG++++ +L WLRQQMGLV QEP+LF
Sbjct: 135 QTVALVGESGSGKSTVIALLQRFYDPDSGEITLDGIEIRQLQLKWLRQQMGLVSQEPVLF 314
Query: 1097 NESIRANIGYGKEGGATEDEIIAAANAANAHSFISNL 1133
N++IRANI YGK G ATE EIIAAA ANAH FIS L
Sbjct: 315 NDTIRANIAYGKGGIATEAEIIAAAELANAHRFISGL 425
Score = 128 bits (322), Expect = 1e-29
Identities = 62/96 (64%), Positives = 79/96 (81%), Gaps = 1/96 (1%)
Frame = +3
Query: 355 TTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFT 414
T ALVG+SGSGKSTVI+LL+RFYDPD+GE+ +DG+ ++ LQL+W+R+Q+GLVSQEP+LF
Sbjct: 138 TVALVGESGSGKSTVIALLQRFYDPDSGEITLDGIEIRQLQLKWLRQQMGLVSQEPVLFN 317
Query: 415 TSIRENIAYGKEG-ATDEEITTAITLANAKKFIDKL 449
+IR NIAYGK G AT+ EI A LANA +FI L
Sbjct: 318 DTIRANIAYGKGGIATEAEIIAAAELANAHRFISGL 425
>TC92328 similar to PIR|T52080|T52080 multi resistance protein [imported] -
Arabidopsis thaliana, partial (18%)
Length = 1133
Score = 136 bits (342), Expect = 6e-32
Identities = 77/238 (32%), Positives = 134/238 (55%)
Frame = +1
Query: 323 GDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAG 382
G IE+ D+ RY + + G S P G +VG++GSGKST+I L R +P G
Sbjct: 100 GTIEIFDLKVRYKENLPL-VLHGVSCTFPGGKNIGIVGRTGSGKSTLIQALFRLIEPADG 276
Query: 383 EVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANA 442
+ ID +N+ + L +R + ++ Q+P LF +IR N+ +E + D++I A+ +
Sbjct: 277 SIHIDNINIFEIGLHDLRSHLSIIPQDPTLFEGTIRGNLDPLEEHS-DKDIWEALDKSQL 453
Query: 443 KKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIV 502
+ I + Q LDT +NG S GQ+Q +++ RA+LK KIL+LDEAT+++D ++ ++
Sbjct: 454 GEIIREKGQKLDTPVIENGDNWSVGQRQLVSLGRALLKQSKILVLDEATASVDTATDNLI 633
Query: 503 QEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLI 560
Q+ + T + +AHR+ T+ ++D + V+ G++ E T L D + +L+
Sbjct: 634 QKIIRTEFKDCTVLTIAHRIPTVIDSDQVLVLSDGRVAEFDTPLRLLEDRSSMFLKLV 807
Score = 125 bits (313), Expect = 1e-28
Identities = 75/244 (30%), Positives = 129/244 (52%)
Frame = +1
Query: 1006 GDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSG 1065
G IE+ + Y + + +S + P K I +VG +GSGKST+I L R +P G
Sbjct: 100 GTIEIFDLKVRYKENLPL-VLHGVSCTFPGGKNIGIVGRTGSGKSTLIQALFRLIEPADG 276
Query: 1066 RILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGATEDEIIAAANAAN 1125
I +D +++ L LR + ++ Q+P LF +IR N+ +E ++ +I A + +
Sbjct: 277 SIHIDNINIFEIGLHDLRSHLSIIPQDPTLFEGTIRGNLDPLEEH--SDKDIWEALDKSQ 450
Query: 1126 AHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESERI 1185
I DT V E G S GQ+Q +++ R +LK KIL+LDEAT+++D ++ +
Sbjct: 451 LGEIIREKGQKLDTPVIENGDNWSVGQRQLVSLGRALLKQSKILVLDEATASVDTATDNL 630
Query: 1186 VQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELMRITDGVYASLVA 1245
+Q+ + + T + +AHR+ T+ +D + V+ +G VAE L+ ++ LV
Sbjct: 631 IQKIIRTEFKDCTVLTIAHRIPTVIDSDQVLVLSDGRVAEFDTPLRLLEDRSSMFLKLVT 810
Query: 1246 LHSS 1249
+SS
Sbjct: 811 EYSS 822
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.136 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,523,628
Number of Sequences: 36976
Number of extensions: 370413
Number of successful extensions: 2262
Number of sequences better than 10.0: 131
Number of HSP's better than 10.0 without gapping: 1984
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2189
length of query: 1250
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1143
effective length of database: 5,058,295
effective search space: 5781631185
effective search space used: 5781631185
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144478.12


