

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144406.8 + phase: 0 /pseudo
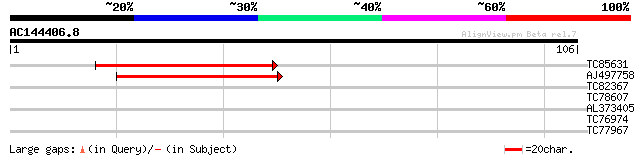
(106 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85631 homologue to SP|Q9BBN9|NU1C_LOTJA NADH-plastoquinone oxi... 48 5e-07
AJ497758 similar to GP|14091074|gb| NADH dehydrogenase subunit 1... 40 2e-04
TC82367 similar to GP|19570986|dbj|BAB86413. diphosphonucleotide... 27 1.6
TC78607 similar to GP|20466746|gb|AAM20690.1 putative protein {A... 25 4.7
AL373405 similar to PIR|F86331|F863 hypothetical protein AAG1254... 25 4.7
TC76974 homologue to GP|9759112|dbj|BAB09597.1 low density lipop... 24 8.0
TC77967 homologue to PIR|T09846|T09846 phosphoenolpyruvate carbo... 24 8.0
>TC85631 homologue to SP|Q9BBN9|NU1C_LOTJA NADH-plastoquinone oxidoreductase
chain 1 chloroplast (EC 1.6.5.3). {Lotus japonicus},
complete
Length = 3706
Score = 48.1 bits (113), Expect = 5e-07
Identities = 22/34 (64%), Positives = 25/34 (72%)
Frame = +3
Query: 17 VMFFISRLAETNRAPFDLPEAEAESVAGYNVEYA 50
++F IS LAE R PFDLPEAE E VAGY EY+
Sbjct: 2628 IVFLISSLAECERLPFDLPEAEEELVAGYQTEYS 2729
>AJ497758 similar to GP|14091074|gb| NADH dehydrogenase subunit 1 {Ostrinia
nubilalis}, partial (26%)
Length = 514
Score = 39.7 bits (91), Expect = 2e-04
Identities = 18/31 (58%), Positives = 21/31 (67%)
Frame = +2
Query: 21 ISRLAETNRAPFDLPEAEAESVAGYNVEYAR 51
IS AE NR PFD E E+E V+G+N EY R
Sbjct: 188 ISCTAELNRTPFDFAEGESELVSGFNTEYRR 280
>TC82367 similar to GP|19570986|dbj|BAB86413. diphosphonucleotide
phosphatase -like protein {Oryza sativa (japonica
cultivar-group)}, partial (3%)
Length = 611
Score = 26.6 bits (57), Expect = 1.6
Identities = 11/26 (42%), Positives = 13/26 (49%)
Frame = +1
Query: 78 GACWGTLHSSFETNECRVGGFCLVIK 103
G C G H FE R+GG+ L K
Sbjct: 400 GVCHGYRHRRFEAERWRLGGYDLTFK 477
>TC78607 similar to GP|20466746|gb|AAM20690.1 putative protein {Arabidopsis
thaliana}, partial (55%)
Length = 1734
Score = 25.0 bits (53), Expect = 4.7
Identities = 23/72 (31%), Positives = 36/72 (49%), Gaps = 2/72 (2%)
Frame = -3
Query: 22 SRLAETNRAPFDLPEAEAESVAGYNVE-YARDAILN-SPLLAEANVPGSRGLILTETRGA 79
+RLA+ N LP AE S+ N++ +A +A SPLL ++VP S L+ G
Sbjct: 601 NRLAKPN-----LPYAEGSSI---NLDKWAENARAKASPLLLASSVPPSLATALSTRSGK 446
Query: 80 CWGTLHSSFETN 91
C + S + +
Sbjct: 445 CSDKIVSQYSNS 410
>AL373405 similar to PIR|F86331|F863 hypothetical protein AAG12548.1
[imported] - Arabidopsis thaliana, partial (64%)
Length = 455
Score = 25.0 bits (53), Expect = 4.7
Identities = 13/40 (32%), Positives = 18/40 (44%), Gaps = 2/40 (5%)
Frame = +2
Query: 22 SRLAETNRAPFDLPEAEAESVAGYNVE--YARDAILNSPL 59
+R++ NRAP D+P G N R LN P+
Sbjct: 119 ARISSANRAPSDVPSGHVAVCVGANYTRFVVRATYLNHPI 238
>TC76974 homologue to GP|9759112|dbj|BAB09597.1 low density lipoprotein
B-like protein {Arabidopsis thaliana}, partial (11%)
Length = 1121
Score = 24.3 bits (51), Expect = 8.0
Identities = 9/23 (39%), Positives = 12/23 (52%)
Frame = -1
Query: 72 ILTETRGACWGTLHSSFETNECR 94
+L +RG CW T F+ CR
Sbjct: 230 LLWPSRGQCWSTDGEVFKPGNCR 162
>TC77967 homologue to PIR|T09846|T09846 phosphoenolpyruvate carboxylase (EC
4.1.1.31) 1 - upland cotton, partial (79%)
Length = 2660
Score = 24.3 bits (51), Expect = 8.0
Identities = 11/39 (28%), Positives = 17/39 (43%)
Frame = +3
Query: 6 IWSGIPLFPVLVMFFISRLAETNRAPFDLPEAEAESVAG 44
IW G+P F V + + R P++ P + S G
Sbjct: 1065 IWKGVPKFLRRVDTALKNIGVNERVPYNAPVIQFSSWMG 1181
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.138 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,292,675
Number of Sequences: 36976
Number of extensions: 34392
Number of successful extensions: 164
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 164
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 164
length of query: 106
length of database: 9,014,727
effective HSP length: 82
effective length of query: 24
effective length of database: 5,982,695
effective search space: 143584680
effective search space used: 143584680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 50 (23.9 bits)
Medicago: description of AC144406.8


