

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144404.22 + phase: 0
(471 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
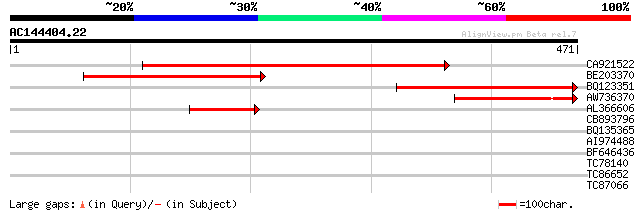
Score E
Sequences producing significant alignments: (bits) Value
CA921522 495 e-140
BE203370 182 3e-46
BQ123351 122 2e-28
AW736370 120 9e-28
AL366606 76 3e-14
CB893796 similar to EGAD|45280|4774 hypothetical protein KIAA022... 30 2.0
BQ135365 weakly similar to GP|17834086|gb| haymaker protein {Mus... 30 2.0
AI974488 similar to PIR|T05841|T058 spliceosome-associated prote... 29 4.4
BF646436 weakly similar to GP|9828634|gb F1N21.5 {Arabidopsis th... 28 5.8
TC78140 similar to PIR|H96781|H96781 unknown protein F22H5.20 [i... 28 9.9
TC86652 similar to GP|19911585|dbj|BAB86896. syringolide-induced... 28 9.9
TC87066 similar to GP|18767124|gb|AAL79277.1 unknown {Saccharomy... 28 9.9
>CA921522
Length = 767
Score = 495 bits (1275), Expect = e-140
Identities = 241/255 (94%), Positives = 245/255 (95%)
Frame = +3
Query: 111 ETDEVKTHLITRGKFLGFSTDFLYERTTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFV 170
ETDEVKTHLITRGKFLGFSTDFLYERTTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFV
Sbjct: 3 ETDEVKTHLITRGKFLGFSTDFLYERTTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFV 182
Query: 171 DIKAIQIFLSKNPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSLLPRTPHFTTN 230
DIKAIQIFLS+NPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITS LPRTP FTTN
Sbjct: 183 DIKAIQIFLSRNPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSHLPRTPRFTTN 362
Query: 231 PENLLWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLARRQF 290
PENLLWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLAR QF
Sbjct: 363 PENLLWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLARHQF 542
Query: 291 GYPMEKKPLSIYLENVYYFNTEDSTGMREQVVRAWHTIRRRDKGQLGKKTGAVHESYTQW 350
GYPME+KPLSIYLENVYY N +DSTGMRE VVRAWHTIRRRDK QLGKKTGA+ SYTQW
Sbjct: 543 GYPMERKPLSIYLENVYYLNADDSTGMREHVVRAWHTIRRRDKDQLGKKTGAISSSYTQW 722
Query: 351 VIDRAVQIRMPYKIS 365
VIDRAVQI MPYKIS
Sbjct: 723 VIDRAVQIGMPYKIS 767
>BE203370
Length = 454
Score = 182 bits (461), Expect = 3e-46
Identities = 92/151 (60%), Positives = 108/151 (70%)
Frame = +1
Query: 62 HCFTFQDYQILPTLEEYSCWINLPVLDKVPFSGLEETPKHSTIAATLHLETDEVKTHLIT 121
HCFTF DYQ+ PTLEEYS + PVLDKVPF+G E PK +TIA LHLET +K L
Sbjct: 1 HCFTFPDYQLAPTLEEYSYLVG*PVLDKVPFTGFEPIPKAATIADALHLETSLIKAKLTI 180
Query: 122 RGKFLGFSTDFLYERTTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFVDIKAIQIFLSK 181
+G T FLY++ + F K+ AF SILALL+YGLVLFP++DN+VDI AIQ+FL+K
Sbjct: 181 KGNLPSLPTKFLYQQASDFAKVDNVDAFYSILALLIYGLVLFPNVDNYVDIHAIQMFLTK 360
Query: 182 NPVPTLLGDTYLSIHRRTQAGRGTILCCAQL 212
NPVPTLL D Y SIH R Q GRG IL CA L
Sbjct: 361 NPVPTLLADMYHSIHDRNQVGRGAILGCAPL 453
>BQ123351
Length = 725
Score = 122 bits (307), Expect = 2e-28
Identities = 61/151 (40%), Positives = 96/151 (63%), Gaps = 1/151 (0%)
Frame = +2
Query: 322 VRAWHTIRRRDKGQLGKKTGAVHESYTQWVIDRAVQIRMPYKISRFVSAITPAPPLPMTF 381
++AW + +R+KGQLG+++ V+ESYT+WVIDRA + +P+ + R S+ T +P LPMT
Sbjct: 11 IQAWRAVHKREKGQLGRRS*LVYESYTKWVIDRAAKNGIPFPLQRLPSSTTLSPSLPMTS 190
Query: 382 DTEKECQEKIAELTRESYMWERKYRKKEQEYDTLKNLLDQQIQANRQEKEENIRLRAALQ 441
T++E Q +AE+TRE W +Y + E E TL+ ++Q+ + +++ I LQ
Sbjct: 191 KTKEEAQYLLAEMTREKDTWRIRYMEAESEIGTLRGQVEQKDHELLKMRQQMIERDDLLQ 370
Query: 442 KKEDFLDK-VCPGRKKRRMDLFDGPHSDFED 471
+K+ L K + ++ MDLFDGP SDFED
Sbjct: 371 EKDKLLKKHITKKQRMDSMDLFDGPDSDFED 463
>AW736370
Length = 488
Score = 120 bits (302), Expect = 9e-28
Identities = 57/102 (55%), Positives = 78/102 (75%)
Frame = -2
Query: 370 AITPAPPLPMTFDTEKECQEKIAELTRESYMWERKYRKKEQEYDTLKNLLDQQIQANRQE 429
AITPAPPLPMTFDTEKE QE++ E RE + W+R+Y+KK+Q+Y+T+ LL+Q+ +R +
Sbjct: 487 AITPAPPLPMTFDTEKEYQERLTEAEREIHRWKREYQKKDQDYETVMGLLEQEAYDSR*K 308
Query: 430 KEENIRLRAALQKKEDFLDKVCPGRKKRRMDLFDGPHSDFED 471
+L +++ + LD++ PGRKKRRMDLFDGPHSDFED
Sbjct: 307 DVIIAKLNERIKENDAALDRI-PGRKKRRMDLFDGPHSDFED 185
>AL366606
Length = 393
Score = 75.9 bits (185), Expect = 3e-14
Identities = 37/58 (63%), Positives = 45/58 (76%)
Frame = -1
Query: 150 NSILALLVYGLVLFPSLDNFVDIKAIQIFLSKNPVPTLLGDTYLSIHRRTQAGRGTIL 207
++I L GL+LFP+LDNFVD+ AI++F SKNPVPTLL DTY +IH RT GRG IL
Sbjct: 384 SAIXLCLSTGLILFPNLDNFVDMNAIEVFHSKNPVPTLLADTYHAIHDRTLKGRGYIL 211
>CB893796 similar to EGAD|45280|4774 hypothetical protein KIAA0226 {Homo
sapiens}, partial (1%)
Length = 660
Score = 30.0 bits (66), Expect = 2.0
Identities = 22/74 (29%), Positives = 36/74 (47%), Gaps = 6/74 (8%)
Frame = +2
Query: 384 EKECQEKIAELTRESY---MWERKYRKKE--QEYDTLKNLLD-QQIQANRQEKEENIRLR 437
+++C+E E TR+S WE KY+KK+ E+D + D EK + L
Sbjct: 164 KRKCEEIRKEYTRKSRRFDQWECKYKKKKGLDEFDQNRGEDDINDPPEVAMEKRVLVELE 343
Query: 438 AALQKKEDFLDKVC 451
L +++D +K C
Sbjct: 344 KRLDEEKDAFEKQC 385
>BQ135365 weakly similar to GP|17834086|gb| haymaker protein {Mus musculus},
partial (7%)
Length = 1147
Score = 30.0 bits (66), Expect = 2.0
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 4/40 (10%)
Frame = +3
Query: 290 FGYPMEKKPLSIYLENV----YYFNTEDSTGMREQVVRAW 325
F +P+ +P S+YL Y +T STG +E VR W
Sbjct: 480 FFWPLPLRPFSLYLYTFLMTHYIKSTNPSTGRKETAVREW 599
>AI974488 similar to PIR|T05841|T058 spliceosome-associated protein homolog
F17L22.120 - Arabidopsis thaliana, partial (5%)
Length = 404
Score = 28.9 bits (63), Expect = 4.4
Identities = 17/49 (34%), Positives = 28/49 (56%)
Frame = -2
Query: 410 QEYDTLKNLLDQQIQANRQEKEENIRLRAALQKKEDFLDKVCPGRKKRR 458
+E + ++N+L QA +E E +LR+ ++EDF D V KKR+
Sbjct: 361 EELEAMENVL----QARYEEAREEEKLRS---QREDFSDMVAENEKKRK 236
>BF646436 weakly similar to GP|9828634|gb F1N21.5 {Arabidopsis thaliana},
partial (11%)
Length = 659
Score = 28.5 bits (62), Expect = 5.8
Identities = 14/49 (28%), Positives = 25/49 (50%)
Frame = +1
Query: 401 WERKYRKKEQEYDTLKNLLDQQIQANRQEKEENIRLRAALQKKEDFLDK 449
W+ K+ LKN+L Q+ + + ++ E RL+ Q ED+L +
Sbjct: 139 WDELDLKRADVEKELKNVLQQKEEILKLQQNEEERLKKEKQATEDYLQR 285
>TC78140 similar to PIR|H96781|H96781 unknown protein F22H5.20 [imported] -
Arabidopsis thaliana, partial (83%)
Length = 1253
Score = 27.7 bits (60), Expect = 9.9
Identities = 12/27 (44%), Positives = 17/27 (62%)
Frame = -1
Query: 206 ILCCAQLLYRWITSLLPRTPHFTTNPE 232
++C LLY + SLLPR PH + P+
Sbjct: 458 LVCHENLLYTFYLSLLPRQPHPSGFPQ 378
>TC86652 similar to GP|19911585|dbj|BAB86896. syringolide-induced protein
13-1-1 {Glycine max}, partial (78%)
Length = 1540
Score = 27.7 bits (60), Expect = 9.9
Identities = 26/75 (34%), Positives = 36/75 (47%), Gaps = 8/75 (10%)
Frame = -2
Query: 73 PTLEEYSCWINLP---VLDKVPFSGLEETPKHSTIAAT-LHLETDEVKTHLITRGKFLGF 128
PT + S + N+P K+P S L + TI T L L ++ +K+ LITRGK F
Sbjct: 1266 PTCNKTSSF*NIPS*IASIKIPSSALSQIL*--TIETTKLELNSETLKSFLITRGKVKAF 1093
Query: 129 S----TDFLYERTTF 139
T FL + F
Sbjct: 1092 DFAICTPFLLSQRQF 1048
>TC87066 similar to GP|18767124|gb|AAL79277.1 unknown {Saccharomyces
cerevisiae}, partial (33%)
Length = 477
Score = 27.7 bits (60), Expect = 9.9
Identities = 10/20 (50%), Positives = 14/20 (70%)
Frame = -3
Query: 47 EGLLNAFVQFYDPGYHCFTF 66
+GLLN F +F+ P +HC F
Sbjct: 469 QGLLNLFSKFFSPFHHCTFF 410
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,997,812
Number of Sequences: 36976
Number of extensions: 217877
Number of successful extensions: 1033
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 1027
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1031
length of query: 471
length of database: 9,014,727
effective HSP length: 100
effective length of query: 371
effective length of database: 5,317,127
effective search space: 1972654117
effective search space used: 1972654117
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144404.22


