

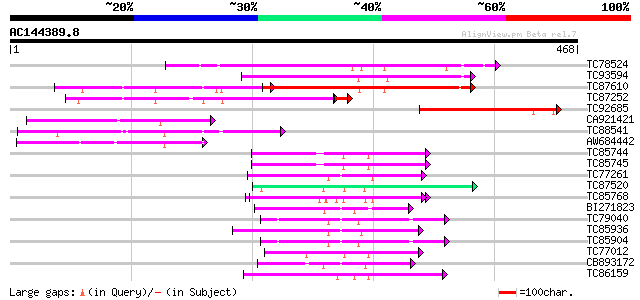
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144389.8 - phase: 0
(468 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78524 similar to PIR|D84561|D84561 probable AAA-type ATPase [i... 215 3e-56
TC93594 weakly similar to GP|20466452|gb|AAM20543.1 unknown prot... 162 3e-40
TC87610 similar to GP|9294102|dbj|BAB01954.1 contains similarity... 157 1e-38
TC87252 similar to GP|11559424|dbj|BAB18781. mitochondrial prote... 128 1e-30
TC92685 similar to GP|10122056|gb|AAG13445.1 hypothetical protei... 127 9e-30
CA921421 weakly similar to GP|18700084|gb| At2g46620/F13A10.15 {... 118 6e-27
TC88541 weakly similar to GP|10176993|dbj|BAB10225. contains sim... 80 2e-15
AW684442 similar to GP|9758309|dbj contains similarity to AAA-ty... 57 1e-08
TC85744 homologue to PIR|T08959|T08959 proteinase homolog F19B15... 57 2e-08
TC85745 homologue to GP|17297987|dbj|BAB78491. 26S proteasome re... 55 4e-08
TC77261 homologue to GP|8777330|dbj|BAA96920.1 26S proteasome AA... 55 6e-08
TC87520 similar to GP|9757998|dbj|BAB08420.1 cell division prote... 52 4e-07
TC85768 homologue to SP|P54774|CC48_SOYBN Cell division cycle pr... 50 1e-06
BI271823 similar to GP|21592745|gb cell division protein FtsH-li... 49 3e-06
TC79040 homologue to PIR|F84674|F84674 probable AAA-type ATPase ... 49 5e-06
TC85936 homologue to GP|13537115|dbj|BAB40755. AtSUG1 {Arabidops... 48 7e-06
TC85904 similar to PIR|F84674|F84674 probable AAA-type ATPase [i... 47 1e-05
TC77012 homologue to SP|P54776|PRSA_LYCES 26S protease regulator... 47 2e-05
CB893172 homologue to PIR|T45642|T45 FtsH metalloproteinase-like... 46 3e-05
TC86159 homologue to GP|21387177|gb|AAM47992.1 26S proteasome AA... 46 3e-05
>TC78524 similar to PIR|D84561|D84561 probable AAA-type ATPase [imported] -
Arabidopsis thaliana, partial (15%)
Length = 1697
Score = 215 bits (548), Expect = 3e-56
Identities = 118/304 (38%), Positives = 184/304 (59%), Gaps = 27/304 (8%)
Frame = +3
Query: 129 FILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYMNASDFGPWRFVPFTHPSTFE 188
F+L + + +++ YI H+ + + I K GN+ L+ + S GPW+ THP++F+
Sbjct: 531 FVLSFDEKHRDKVMEKYIPHVLSTYEAI-KAGNKTLKIHSMQS--GPWKQSDLTHPASFD 701
Query: 189 TITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSY 248
++ M+ DLKN + DL+ FL+ K+ Y ++G+ WKR +LLYG GTGKSS IAAMA +L +
Sbjct: 702 SLAMDPDLKNSIIDDLDRFLRRKKLYKKVGKPWKRGYLLYGPPGTGKSSLIAAMAKYLKF 881
Query: 249 DVYYIDLSRISTDSDLKSILLQTAPKSIIVVED-------LDRYLTEK------------ 289
DVY +DLS + ++S+L + +T+ +SIIV ED LDR +K
Sbjct: 882 DVYDLDLSSVFSNSELMRAMRETSNRSIIVFEDIDCNSEVLDRAKPDKFPDMDFLDGIKM 1061
Query: 290 ----SSTTVTSSGILNFMDGIWS--GEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLC 343
T SG+LN+MDG+WS GEER+++FT N K+ VDP LLRPGR+D+HIH
Sbjct: 1062GKNMPPRKFTLSGLLNYMDGLWSSCGEERILIFTTNHKDKVDPALLRPGRMDMHIHLSFL 1241
Query: 344 DFSSFKTLASNYLGVK--DHKLFPQVQEIFENGASLSPAEIGELMIANRNSPSRAIKTVI 401
+F+ LA+NYL ++ H LF Q++E+ E +SPA + E ++ + + P A+ ++
Sbjct: 1242KAKAFRILAANYLDIEGNHHSLFEQIEELLEK-VDVSPAVVAEYLLRSED-PDVALGALV 1415
Query: 402 TALK 405
L+
Sbjct: 1416KFLQ 1427
>TC93594 weakly similar to GP|20466452|gb|AAM20543.1 unknown protein
{Arabidopsis thaliana}, partial (40%)
Length = 754
Score = 162 bits (409), Expect = 3e-40
Identities = 85/234 (36%), Positives = 129/234 (54%), Gaps = 41/234 (17%)
Frame = +1
Query: 192 METDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVY 251
M K + DL +F K K+YY ++G+ WKR +LLYG GTGKSS +AA+ANFL YD+Y
Sbjct: 4 MSPKKKKEIVDDLVTFSKAKEYYAKIGKPWKRGYLLYGPPGTGKSSLVAAIANFLKYDIY 183
Query: 252 YIDLSRISTDSDLKSILLQTAPKSIIVVEDLDRYL------------------------- 286
I+L+ + +++L+ +L+ KS++V+ED+D L
Sbjct: 184 DIELTNVKNNAELRKLLIGITSKSVVVIEDIDCSLDLTGQRKTDSENDKEKEEKNEEVNQ 363
Query: 287 --------------TEKSSTTVTSSGILNFMDGIWSGE--ERVMVFTMNSKENVDPNLLR 330
+ ++ VT SG+LNF+DGIWS ER+++FT N E +D L+R
Sbjct: 364 VAAASLQGLQAADKEKNKASQVTLSGLLNFIDGIWSASTGERLIIFTTNYVEKLDQALIR 543
Query: 331 PGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQVQEIFENGASLSPAEIGE 384
GR+D+HI F FK LA NYL ++ H LF +Q + E +++PA++ E
Sbjct: 544 RGRMDMHIELSYWGFDGFKMLAMNYLSIESHPLFETIQRLLEE-TNMTPADVAE 702
>TC87610 similar to GP|9294102|dbj|BAB01954.1 contains similarity to
AAA-type ATPase~gene_id:T20D4.3 {Arabidopsis thaliana},
partial (53%)
Length = 1765
Score = 157 bits (396), Expect = 1e-38
Identities = 83/208 (39%), Positives = 128/208 (60%), Gaps = 32/208 (15%)
Frame = +3
Query: 209 KGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSIL 268
KGK +L + WKR +LL+G GTGKS+ I+A+ANF++YDVY ++L+ + +++LK +L
Sbjct: 756 KGKSIMLKLEKAWKRGYLLFGPPGTGKSTMISAIANFMNYDVYDLELTIVKDNNELKRLL 935
Query: 269 LQTAPKSIIVVEDLDRYLT-----------------------------EKSSTTVTSSGI 299
++T+ KSIIV+ED+D L EK+ + VT SG+
Sbjct: 936 IETSSKSIIVIEDIDCSLDLTGQRKKKKEKDDVENDEKKDPIKKAEKEEKNESKVTLSGL 1115
Query: 300 LNFMDGIWS--GEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLG 357
LNF+DGIWS G ER+++FT N + +DP L+R GR+D HI C + +FK LA NYL
Sbjct: 1116LNFIDGIWSACGSERIIIFTTNFVDKLDPALIRRGRMDKHIEMSYCSYQAFKVLARNYLD 1295
Query: 358 VKDH-KLFPQVQEIFENGASLSPAEIGE 384
V+ H LFP ++++ +++PA++ E
Sbjct: 1296VETHDDLFPIIEKLL-GETNMTPADVAE 1376
Score = 81.3 bits (199), Expect = 7e-16
Identities = 59/211 (27%), Positives = 95/211 (44%), Gaps = 29/211 (13%)
Frame = +2
Query: 38 LWRIIEDWFHVYQVFHVPELN--------DNMQHNTLYRKLSLYFHSLPSLQNSQLNNLV 89
L R + + H + F P + DN++HN Y + Y + S + +L V
Sbjct: 161 LRRYLRKYTHKFTNFMYPYIKITFYEKSGDNLKHNKTYTTIQTYLSANSSQRARRLKAEV 340
Query: 90 TSNTNQNDVVLTLAPNQTIHDHFLGATVSW--------------FNQTQPNRTFILRIRK 135
++ QN +VL++ NQ I D F G V W + + R L K
Sbjct: 341 IKDS-QNPLVLSMDDNQEITDEFNGVKVWWSANHITSRTQSFSIYPSSDEKRFLTLTFHK 517
Query: 136 FDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYMN--ASDF-----GPWRFVPFTHPSTFE 188
++ I +YIQH+ I + NR L+ Y N ++D+ W F HP++FE
Sbjct: 518 RHRELITTSYIQHVLEQGKAITMK-NRQLKIYTNNPSNDWFRYRSTKWSHTTFEHPASFE 694
Query: 189 TITMETDLKNRVKSDLESFLKGKQYYHRLGR 219
T+ +E K + +DL F KGK+YY ++G+
Sbjct: 695 TLALEPKKKEEILNDLVKFKKGKEYYAKVGK 787
>TC87252 similar to GP|11559424|dbj|BAB18781. mitochondrial protein-like
protein {Cucumis sativus}, partial (72%)
Length = 968
Score = 128 bits (322), Expect(2) = 1e-30
Identities = 79/251 (31%), Positives = 131/251 (51%), Gaps = 26/251 (10%)
Frame = +2
Query: 47 HVYQVFHVP--ELN-DNMQHNTLYRKLSLYFHSLPSLQNSQLNNLVTSNTNQNDVVLTLA 103
++Y H+ EL+ + ++ + Y+ + Y S + +L V ++ QN +VL++
Sbjct: 185 YMYPYIHIKFHELSGERLKQSETYKIIQTYLSDNSSQRARRLKAEVVKDS-QNPLVLSMD 361
Query: 104 PNQTIHDHFLGATVSW--------------FNQTQPNRTFILRIRKFDKQRILRAYIQHI 149
N+ I D F G V W + + R L K ++ I +YIQH
Sbjct: 362 DNEEIIDEFNGVKVWWTANYTTSKSQSFSYYPTSDEKRFLTLTFHKKHREVITTSYIQH- 538
Query: 150 HAVVDEIEK--QGNRDLRFYMNASDFG-------PWRFVPFTHPSTFETITMETDLKNRV 200
V+DE + NR L+ Y N W F HP+ F T+ ME + K +
Sbjct: 539 --VLDEGKSIMSKNRQLKLYTNNPSSNWWGYRSKKWNHTTFEHPARFGTLAMEPEKKQEI 712
Query: 201 KSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRIST 260
+DL F KGK+YY ++G+ WKR +LLYG GTGKS+ I+A+AN+++YDVY ++L+ +
Sbjct: 713 LNDLLKFKKGKEYYAKVGKAWKRGYLLYGPPGTGKSTMISAIANYMNYDVYDLELTTVKD 892
Query: 261 DSDLKSILLQT 271
+++LK +L++T
Sbjct: 893 NNELKRLLIET 925
Score = 22.7 bits (47), Expect(2) = 1e-30
Identities = 9/15 (60%), Positives = 13/15 (86%)
Frame = +3
Query: 269 LQTAPKSIIVVEDLD 283
L+ + KSIIV+ED+D
Sbjct: 918 LKRSSKSIIVIEDID 962
>TC92685 similar to GP|10122056|gb|AAG13445.1 hypothetical protein {Oryza
sativa (japonica cultivar-group)}, partial (12%)
Length = 665
Score = 127 bits (319), Expect = 9e-30
Identities = 69/131 (52%), Positives = 88/131 (66%), Gaps = 14/131 (10%)
Frame = +1
Query: 339 HFPLCDFSSFKTLASNYLGVKDHKLFPQVQEIF-ENGASLSPAEIGELMIANRNSPSRAI 397
HFPLCDFS+FK LAS+YLG+K+HKLFPQV+E+F + GA LSPAE+GE+MI+NRNSPSRA+
Sbjct: 1 HFPLCDFSTFKILASSYLGLKEHKLFPQVEEVFYQTGARLSPAEVGEIMISNRNSPSRAL 180
Query: 398 KTVITALKTDGDGRGCGFIERRIGNEGDGVDEGA-----------RDTRKLYGFFRLKGP 446
KTVITA++ +G G G + V++ + R+ RKLYG RL
Sbjct: 181 KTVITAMQVQSNGSGQRLSHSGSGRSSEEVNDTSAVICRESVHTVREFRKLYGLLRLGSR 360
Query: 447 R--KSSSSPNS 455
R K S PNS
Sbjct: 361 RKEKDESVPNS 393
>CA921421 weakly similar to GP|18700084|gb| At2g46620/F13A10.15 {Arabidopsis
thaliana}, partial (11%)
Length = 582
Score = 118 bits (295), Expect = 6e-27
Identities = 59/159 (37%), Positives = 93/159 (58%), Gaps = 3/159 (1%)
Frame = +3
Query: 15 ILLFSIYAIHMIFETGLIHESTKLWRIIEDWFHVYQVFHVPELNDNMQHNTLYRKLSLYF 74
I +FS + +T +H W +E+ H++Q F +P + N + N LYRK+ Y
Sbjct: 93 IFVFSYLILRFFRKTSALHFLKHWWLSLENRLHLHQSFKIPLYDHNFRENQLYRKILTYL 272
Query: 75 HSLPSLQNSQLNNLVTSNTNQNDVVLTLAPNQTIHDHFLGATVSWFNQT---QPNRTFIL 131
SLPS+Q++ NL S N +D+ L L NQ +HD FLGA +SW N T +L
Sbjct: 273 DSLPSVQDADFTNLF-SGPNPSDIFLHLDANQIVHDTFLGAKLSWTNNTVAGDSASALVL 449
Query: 132 RIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYMNA 170
R++K DK+R+ + Y QHI +V DE+E++ +D++ +MN+
Sbjct: 450 RMKKKDKRRVFQQYFQHILSVADELEQRRKKDIKLFMNS 566
>TC88541 weakly similar to GP|10176993|dbj|BAB10225. contains similarity to
AAA-type ATPase~gene_id:MYH19.21 {Arabidopsis thaliana},
partial (26%)
Length = 854
Score = 80.1 bits (196), Expect = 2e-15
Identities = 61/240 (25%), Positives = 107/240 (44%), Gaps = 19/240 (7%)
Frame = +1
Query: 7 TPILLKIAILLFSIYAIHMIFETGLIHESTKL-----WRIIEDWFHVYQVFHVPELNDNM 61
T + +I ++ S+ I IF+ ++ L R++ + Q+ + +
Sbjct: 148 TEMFAQIGSIIASLMFIWAIFQQYFPYQLRNLIDKYSQRLVTFIYPYIQITFHEFTGERL 327
Query: 62 QHNTLYRKLSLYFHSLPSLQNSQLNNLVTSNTNQNDVVLTLAPNQTIHDHFLGATVSWFN 121
+ Y + Y S S Q +L + N NQ+ ++L++ + I D F G + W +
Sbjct: 328 MRSEAYSSIENYLSSKASTQAKRLKGDIAKN-NQS-LILSMDDKEEICDEFNGMKLWWAS 501
Query: 122 QTQPN--------------RTFILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFY 167
+ + R + L K ++ IL Y+ H+ I+ + NR + Y
Sbjct: 502 GKKASNSNSISLHQNIDEKRYYKLTFHKHNRDVILGKYLSHVLKEGKAIQVK-NRQRKLY 678
Query: 168 MNASDFGPWRFVPFTHPSTFETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLL 227
N+ W V F HPSTFET+ M+ + K + DL +F K ++Y R+GR WKR +LL
Sbjct: 679 TNSGSH--WSHVVFEHPSTFETLAMDLEKKEMIIDDLITFSKAGEFYARIGRAWKRGYLL 852
>AW684442 similar to GP|9758309|dbj contains similarity to AAA-type
ATPase~gene_id:MUA2.5 {Arabidopsis thaliana}, partial
(33%)
Length = 600
Score = 57.4 bits (137), Expect = 1e-08
Identities = 48/172 (27%), Positives = 78/172 (44%), Gaps = 14/172 (8%)
Frame = +2
Query: 6 YTPILLKIAILLFSIYAIHMIFETGLIHESTKLWRIIEDWFHVYQVFHVPELNDNMQHNT 65
+T + + + F + +F L S KL+ + + F Y F + E+ D + N
Sbjct: 89 WTSLASILGVFAFFQTILQTVFPPELRFASAKLFNKLFNCFSSYCYFEITEI-DGVNTNE 265
Query: 66 LYRKLSLYFHSLPSLQNSQLNNLVTSNTNQNDVVLTLAPNQTIHDHFLGATVSWFNQ-TQ 124
LY + LY S S+ ++L+ +T N + LA N +I D F G V W + TQ
Sbjct: 266 LYNAVQLYLSSSVSITGNRLS--LTRAVNSSAFTFGLANNDSIIDTFNGVNVVWEHVVTQ 439
Query: 125 PN-------------RTFILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRD 163
N R F LRI+K DKQ + +Y+ +I +I ++ N+D
Sbjct: 440 RNSQTFSWRPLPDEKRGFTLRIKKKDKQLLXNSYLDYIMEKASDIRRK-NQD 592
>TC85744 homologue to PIR|T08959|T08959 proteinase homolog F19B15.70 -
Arabidopsis thaliana, complete
Length = 1629
Score = 56.6 bits (135), Expect = 2e-08
Identities = 47/170 (27%), Positives = 73/170 (42%), Gaps = 22/170 (12%)
Frame = +2
Query: 200 VKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRIS 259
+K +E L + Y +G + +LYG GTGK+ A+AN S +
Sbjct: 689 IKEAVELPLTHPELYEDIGIKPPKGVILYGEPGTGKTLLAKAVANSTSATFLRV------ 850
Query: 260 TDSDLKSILLQTAPK--------------SIIVVEDLDRYLTEKSSTTV--------TSS 297
S+L L PK SI+ ++++D T++ T
Sbjct: 851 VGSELIQKYLGDGPKLVRELFRVADDLSPSIVFIDEIDAVGTKRYDAHSGGEREIQRTML 1030
Query: 298 GILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSS 347
+LN +DG S + ++ N E++DP LLRPGR+D I FPL D +
Sbjct: 1031ELLNQLDGFDSRGDVKVILATNRIESLDPALLRPGRIDRKIEFPLPDIKT 1180
>TC85745 homologue to GP|17297987|dbj|BAB78491. 26S proteasome regulatory
particle triple-A ATPase subunit2b {Oryza sativa
(japonica cultivar-group), partial (98%)
Length = 1696
Score = 55.5 bits (132), Expect = 4e-08
Identities = 46/170 (27%), Positives = 73/170 (42%), Gaps = 22/170 (12%)
Frame = +3
Query: 200 VKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRIS 259
+K +E L + Y +G + +LYG GTGK+ A+AN S +
Sbjct: 687 IKEAVELPLTHPELYEDIGIKPPKGVILYGEPGTGKTLLAKAVANSTSATFLRV------ 848
Query: 260 TDSDLKSILLQTAPK--------------SIIVVEDLDRYLTEKSSTTV--------TSS 297
S+L L PK +I+ ++++D T++ T
Sbjct: 849 VGSELIQKYLGDGPKLVRELFRVADDLSPAIVFIDEIDAVGTKRYDAHSGGEREIQRTML 1028
Query: 298 GILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSS 347
+LN +DG S + ++ N E++DP LLRPGR+D I FPL D +
Sbjct: 1029ELLNQLDGFDSRGDVKVILATNRIESLDPALLRPGRIDRKIEFPLPDIKT 1178
>TC77261 homologue to GP|8777330|dbj|BAA96920.1 26S proteasome AAA-ATPase
subunit RPT3 {Arabidopsis thaliana}, partial (94%)
Length = 1575
Score = 55.1 bits (131), Expect = 6e-08
Identities = 45/165 (27%), Positives = 71/165 (42%), Gaps = 17/165 (10%)
Frame = +2
Query: 197 KNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLS 256
K ++ +E L + Y ++G R LLYG GTGK+ A+AN + + S
Sbjct: 602 KQEIREAVELPLTHHELYKQIGIDPPRGVLLYGPPGTGKTMLAKAVANHTTAAFIRVVGS 781
Query: 257 RISTD---------SDLKSILLQTAPKSIIVVEDLDRYLTEKSSTTVTSSG--------I 299
D+ + + AP +II ++++D T + + +
Sbjct: 782 EFVQKYLGEGPRMVRDVFRLAKENAP-AIIFIDEVDAIATARFDAQTGADREVQRILMEL 958
Query: 300 LNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCD 344
LN MDG ++ N + +DP LLRPGR+D I FPL D
Sbjct: 959 LNQMDGFDQTVNVKVIMATNRADTLDPALLRPGRLDRKIEFPLPD 1093
>TC87520 similar to GP|9757998|dbj|BAB08420.1 cell division protein FtsH
protease-like {Arabidopsis thaliana}, partial (53%)
Length = 1667
Score = 52.4 bits (124), Expect = 4e-07
Identities = 52/201 (25%), Positives = 82/201 (39%), Gaps = 15/201 (7%)
Frame = +2
Query: 201 KSDLES---FLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSR 257
K +LE +LK + RLG + LL G+ GTGK+ A+A +Y S
Sbjct: 5 KQELEEVVEYLKNPAKFTRLGGKLPKGILLTGAPGTGKTLLAKAIAGEAGVPFFYRAGSE 184
Query: 258 -----ISTDSDLKSILLQTAPKS---IIVVEDLDRYLTEKSS----TTVTSSGILNFMDG 305
+ + L Q A K II ++++D + + T T +L MDG
Sbjct: 185 FEEMFVGVGARRVRSLFQAAKKKAPCIIFIDEIDAVGSTRKQWEGHTKKTLHQLLVEMDG 364
Query: 306 IWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLFP 365
E +++ N + +DP L RPGR D HI P D + + YL K
Sbjct: 365 FEQNEGIILMAATNLPDILDPALTRPGRFDRHIVVPNPDVRGRQEILELYLHDKPTADNV 544
Query: 366 QVQEIFENGASLSPAEIGELM 386
++ I + A++ L+
Sbjct: 545 DIKAIARGTPGFNGADLANLV 607
>TC85768 homologue to SP|P54774|CC48_SOYBN Cell division cycle protein 48
homolog (Valosin containing protein homolog) (VCP).
[Soybean], partial (95%)
Length = 2785
Score = 50.4 bits (119), Expect = 1e-06
Identities = 38/169 (22%), Positives = 79/169 (46%), Gaps = 16/169 (9%)
Frame = +2
Query: 195 DLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYID 254
++K ++ ++ ++ + + + G + L YG G GK+ A+AN + I
Sbjct: 1556 NVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLAKAIANECQANFISIK 1735
Query: 255 LSRIST------DSDLKSIL--LQTAPKSIIVVEDLDRYLTEKSSTTVTSSG-------- 298
+ T +++++ I + + ++ ++LD T++ S+ + G
Sbjct: 1736 GPELLTMWFGESEANVREIFDKARGSAPCVLFFDELDSIATQRGSSVGDAGGAADRVLNQ 1915
Query: 299 ILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSS 347
+L MDG+ + + ++ N + +DP LLRPGR+D I+ PL D S
Sbjct: 1916 LLTEMDGMSAKKTVFIIGATNRPDIIDPALLRPGRLDQLIYIPLPDEDS 2062
Score = 45.1 bits (105), Expect = 6e-05
Identities = 39/159 (24%), Positives = 75/159 (46%), Gaps = 13/159 (8%)
Frame = +2
Query: 199 RVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYID---- 254
+++ +E L+ Q + +G + LLYG G+GK+ A+AN + I+
Sbjct: 749 QIRELVELPLRHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVANETGAFFFCINGPEI 928
Query: 255 LSRIS--TDSDLKSILLQTAPK--SIIVVEDLDRYLTEKSST-----TVTSSGILNFMDG 305
+S+++ ++S+L+ + SII ++++D ++ T S +L MDG
Sbjct: 929 MSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKTHGEVERRIVSQLLTLMDG 1108
Query: 306 IWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCD 344
+ S +++ N ++DP L R GR D I + D
Sbjct: 1109LKSRAHVIVMGATNRPNSIDPALRRFGRFDREIDIGVPD 1225
>BI271823 similar to GP|21592745|gb cell division protein FtsH-like protein
{Arabidopsis thaliana}, partial (33%)
Length = 714
Score = 49.3 bits (116), Expect = 3e-06
Identities = 45/147 (30%), Positives = 64/147 (42%), Gaps = 16/147 (10%)
Frame = +3
Query: 203 DLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRI---- 258
++ S L+G Y +LG R LL G GTGK+ A+A + + S
Sbjct: 141 EIVSCLQGDINYQKLGAKLPRGVLLVGPPGTGKTLLARAVAGEAGVPFFTVSASEFVEMF 320
Query: 259 -----STDSDLKSILLQTAPKSIIVVEDLD-------RYLTEKSSTTVTSSGILNFMDGI 306
+ DL S + AP SII +++LD R E+ T+ +L MDG
Sbjct: 321 VGRGAARIRDLFSRARKFAP-SIIFIDELDAVGGKRGRGFNEERDQTLNQ--LLTEMDGF 491
Query: 307 WSGEERVMVFTMNSKENVDPNLLRPGR 333
S V++ N E +DP L RPGR
Sbjct: 492 ESEIRVVVIAATNRPEALDPALCRPGR 572
>TC79040 homologue to PIR|F84674|F84674 probable AAA-type ATPase [imported]
- Arabidopsis thaliana, partial (74%)
Length = 1193
Score = 48.5 bits (114), Expect = 5e-06
Identities = 47/172 (27%), Positives = 80/172 (46%), Gaps = 16/172 (9%)
Frame = +3
Query: 208 LKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTD------ 261
+K Q++ R W R+FLLYG GTGKS A+A + I S + +
Sbjct: 120 VKFPQFFTGKRRPW-RAFLLYGPPGTGKSYLAKAVATEADSTFFSISSSDLVSKWMGESE 296
Query: 262 ---SDLKSILLQTAPKSIIVVEDLDRYL------TEKSSTTVTSSGILNFMDGIWSGEER 312
S+L + ++AP SII V+++D E ++ + +L M G+ + +++
Sbjct: 297 KLVSNLFQMARESAP-SIIFVDEIDSLCGQRGEGNESEASRRIKTELLVQMQGVGNNDQK 473
Query: 313 VMVF-TMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKL 363
V+V N+ +D + R R D I+ PL D + + + +LG H L
Sbjct: 474 VLVLAATNTPYALDQAIRR--RFDKRIYIPLPDLKARQHMFKVHLGDTPHNL 623
>TC85936 homologue to GP|13537115|dbj|BAB40755. AtSUG1 {Arabidopsis
thaliana}, partial (98%)
Length = 1633
Score = 48.1 bits (113), Expect = 7e-06
Identities = 44/175 (25%), Positives = 75/175 (42%), Gaps = 18/175 (10%)
Frame = +1
Query: 185 STFETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMAN 244
ST++ I +K +E +K + + LG + LLYG GTGK+ A+A+
Sbjct: 613 STYDMIGGLDQQIKEIKEVIELPIKHPELFESLGIAQPKGVLLYGPPGTGKTLLARAVAH 792
Query: 245 FLSYDVYYIDLSRISTD---------SDLKSILLQTAPKSIIVVEDLDRYLTEK------ 289
+ S + +L + + AP SII ++++D + +
Sbjct: 793 HTDCTFIRVSGSELVQKYIGEGSRMVRELFVMAREHAP-SIIFMDEIDSIGSARMESGSG 969
Query: 290 ---SSTTVTSSGILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFP 341
S T +LN +DG + + ++ N + +D LLRPGR+D I FP
Sbjct: 970 NGDSEVQRTMLELLNQLDGFEASNKIKVLMATNRIDILDQALLRPGRIDRKIEFP 1134
>TC85904 similar to PIR|F84674|F84674 probable AAA-type ATPase [imported] -
Arabidopsis thaliana, complete
Length = 1793
Score = 47.4 bits (111), Expect = 1e-05
Identities = 46/172 (26%), Positives = 79/172 (45%), Gaps = 16/172 (9%)
Frame = +2
Query: 208 LKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTD------ 261
+K Q++ R W R+FLLYG GTGKS A+A + + S + +
Sbjct: 638 VKFPQFFTGKRRPW-RAFLLYGPPGTGKSYLAKAVATEADSTFFSVSSSDLVSKWMGESE 814
Query: 262 ---SDLKSILLQTAPKSIIVVEDLDRYL------TEKSSTTVTSSGILNFMDGIWSGEER 312
S+L + ++AP SII V+++D E ++ + +L M G+ +++
Sbjct: 815 KLVSNLFEMARESAP-SIIFVDEIDSLCGTRGEGNESEASRRIKTELLVQMQGVGHNDQK 991
Query: 313 VMVF-TMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKL 363
V+V N+ +D + R R D I+ PL D + + + +LG H L
Sbjct: 992 VLVLAATNTPYALDQAIRR--RFDKRIYIPLPDLKARQHMFKVHLGDTPHNL 1141
>TC77012 homologue to SP|P54776|PRSA_LYCES 26S protease regulatory subunit
6A homolog (TAT-binding protein homolog 1) (TBP-1)
(Mg(2+), complete
Length = 1793
Score = 47.0 bits (110), Expect = 2e-05
Identities = 41/147 (27%), Positives = 65/147 (43%), Gaps = 16/147 (10%)
Frame = +1
Query: 211 KQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMA-----NFLSYDVYYIDLSRISTDSDLK 265
K+ + +LG + LLYG GTGK+ A A FL + I + L
Sbjct: 709 KERFQKLGIRPPKGVLLYGPPGTGKTLMARACAAQTNATFLKLAGPQLVQMFIGDGAKLV 888
Query: 266 SILLQTAPKS---IIVVEDLDRYLTEKSSTTV--------TSSGILNFMDGIWSGEERVM 314
Q A + II ++++D T++ + V T +LN +DG S + +
Sbjct: 889 RDAFQLAKEKSPCIIFIDEIDAIGTKRFDSEVSGDREVQRTMLELLNQLDGFSSDDRIKV 1068
Query: 315 VFTMNSKENVDPNLLRPGRVDVHIHFP 341
+ N + +DP L+R GR+D I FP
Sbjct: 1069IAATNRADILDPALMRSGRLDRKIEFP 1149
>CB893172 homologue to PIR|T45642|T45 FtsH metalloproteinase-like protein -
Arabidopsis thaliana, partial (25%)
Length = 661
Score = 46.2 bits (108), Expect = 3e-05
Identities = 44/151 (29%), Positives = 63/151 (41%), Gaps = 20/151 (13%)
Frame = +3
Query: 205 ESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSR------- 257
+ FL+ Y RLG R LL G GTGK+ A+A DV +I S
Sbjct: 33 QEFLRNPDRYARLGARPPRGVLLVGLPGTGKTLLAKAVAG--EADVPFISCSASEFVELY 206
Query: 258 ----ISTDSDLKSILLQTAPKSIIVVEDLDRYLTEKSST---------TVTSSGILNFMD 304
S DL + + AP SII ++++D + T + +L MD
Sbjct: 207 VGMGASRVRDLFARAKKEAP-SIIFIDEIDAVAKSRDGKFRIVGNDEREQTLNQLLTEMD 383
Query: 305 GIWSGEERVMVFTMNSKENVDPNLLRPGRVD 335
G S +++ N + +DP L RPGR D
Sbjct: 384 GFDSNSPVIVLAATNRADVLDPALRRPGRFD 476
>TC86159 homologue to GP|21387177|gb|AAM47992.1 26S proteasome AAA-ATPase
subunit RPT4a-like protein {Arabidopsis thaliana},
complete
Length = 1640
Score = 45.8 bits (107), Expect = 3e-05
Identities = 48/185 (25%), Positives = 81/185 (42%), Gaps = 17/185 (9%)
Frame = +2
Query: 194 TDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYI 253
+D ++ +E L + + R+G + LLYG GTGK+ A+A+ + + +
Sbjct: 503 SDQIRELRESIELPLMNPELFLRVGIKPPKGVLLYGPPGTGKTLLARAIASNIDANFLKV 682
Query: 254 DLSRISTDSDLKSILL--------QTAPKSIIVVEDLD----RYLTEKSSTTV----TSS 297
S I +S L + II ++++D R +E +S T
Sbjct: 683 VSSAIIDKYIGESARLIREMFGYARDHQPCIIFMDEIDAIGGRRFSEGTSADREIQRTLM 862
Query: 298 GILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPL-CDFSSFKTLASNYL 356
+LN +DG + M+ N + +DP LLRPGR+D I PL + S + L +
Sbjct: 863 ELLNQLDGFDQLGKVKMIMATNRPDVLDPALLRPGRLDRKIEIPLPNEQSRMEILKIHAA 1042
Query: 357 GVKDH 361
G+ H
Sbjct: 1043GIAKH 1057
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,459,027
Number of Sequences: 36976
Number of extensions: 202420
Number of successful extensions: 1296
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 1266
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1275
length of query: 468
length of database: 9,014,727
effective HSP length: 100
effective length of query: 368
effective length of database: 5,317,127
effective search space: 1956702736
effective search space used: 1956702736
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144389.8


