

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144375.8 + phase: 0
(396 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
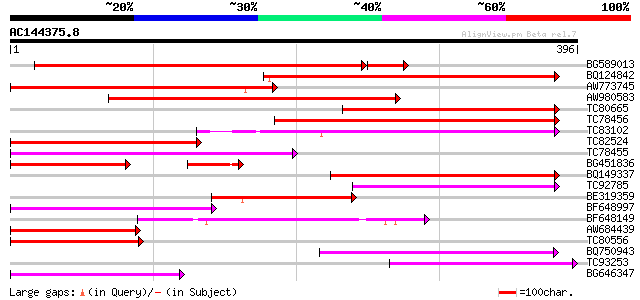
Score E
Sequences producing significant alignments: (bits) Value
BG589013 weakly similar to GP|20805025|db contains EST D15170(C0... 369 e-107
BQ124842 weakly similar to GP|19310633|gb unknown protein {Arabi... 184 4e-47
AW773745 weakly similar to GP|19310633|gb unknown protein {Arabi... 179 2e-45
AW980583 weakly similar to GP|16797803|db hypothetical membrane ... 164 7e-41
TC80665 weakly similar to GP|19310633|gb|AAL85047.1 unknown prot... 162 2e-40
TC78456 similar to GP|11994404|dbj|BAB02363. gb|AAC28507.1~gene_... 155 3e-38
TC83102 similar to GP|11994404|dbj|BAB02363. gb|AAC28507.1~gene_... 136 1e-32
TC82524 similar to GP|19310633|gb|AAL85047.1 unknown protein {Ar... 129 3e-30
TC78455 similar to GP|11994404|dbj|BAB02363. gb|AAC28507.1~gene_... 127 7e-30
BG451836 weakly similar to PIR|A96736|A96 hypothetical protein F... 92 2e-28
BQ149337 weakly similar to GP|13272459|gb unknown protein {Arabi... 117 6e-27
TC92785 weakly similar to GP|13937234|gb|AAK50109.1 AT5g38030/F1... 111 6e-25
BE319359 similar to GP|19310633|gb| unknown protein {Arabidopsis... 109 2e-24
BF648997 weakly similar to GP|13384114|gb aberrant lateral root ... 108 3e-24
BF648149 similar to PIR|T05135|T05 hypothetical protein F7H19.22... 106 1e-23
AW684439 weakly similar to PIR|F86285|F86 hypothetical protein A... 102 3e-22
TC80556 weakly similar to PIR|F86285|F86285 hypothetical protein... 102 3e-22
BQ750943 weakly similar to GP|21627817|emb hypothetical protein ... 100 2e-21
TC93253 weakly similar to GP|14030731|gb|AAK53040.1 AT5g65380/MN... 86 2e-17
BG646347 similar to PIR|T05135|T05 hypothetical protein F7H19.22... 84 7e-17
>BG589013 weakly similar to GP|20805025|db contains EST D15170(C0202)~unknown
protein {Oryza sativa (japonica cultivar-group)},
partial (42%)
Length = 786
Score = 369 bits (946), Expect(2) = e-107
Identities = 171/232 (73%), Positives = 203/232 (86%)
Frame = +1
Query: 18 FSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIAQAAREYCICLIPALFGH 77
+ K+GNY C A++TL +VCFPISL+WIF DK+L+ F QD ++ AREYCI LIPALFG+
Sbjct: 1 YGKLGNYTCCAILTLTVVCFPISLVWIFTDKILMFFSQDPGMSHVAREYCIYLIPALFGY 180
Query: 78 AVLQSLIRYFQIQSMIFPMVFSSIVILCLHVPICWCLVFKFGLGHVGAAFAIGIAYWLNV 137
A+LQ+LIRYFQ Q MIFPMVFSS+ L LH+PICW LVFK GLGH+GAA AIGI+YWLNV
Sbjct: 181 ALLQALIRYFQTQGMIFPMVFSSVSALFLHIPICWILVFKLGLGHIGAALAIGISYWLNV 360
Query: 138 IWLGIYMKYSPACEKTKIVFSYNSLLYIAEFCQFAIPSGLMFCLEWWSFEILTIVAGLLP 197
IWL +Y+KYSP+CEKTKIVFS ++L + EFC++AIPSGLMFC EWWSFEIL ++AGLLP
Sbjct: 361 IWLWVYIKYSPSCEKTKIVFSTHALHNLPEFCKYAIPSGLMFCFEWWSFEILILIAGLLP 540
Query: 198 NSQLETSVLSVCLSTTTLHYFIPHAIGASASTRVSNELGAGNPRAAKGAVRV 249
N QLETSVLSVCL+TT+LH+FIP+AIGASASTRVSNELGAGNP+ AKGAVRV
Sbjct: 541 NPQLETSVLSVCLNTTSLHFFIPYAIGASASTRVSNELGAGNPKTAKGAVRV 696
Score = 39.7 bits (91), Expect(2) = e-107
Identities = 20/28 (71%), Positives = 22/28 (78%)
Frame = +2
Query: 251 VIIGIAEAVIVSTLFLCFRNIIGNAYSN 278
VIIG A+IVST FL FRNI+G AYSN
Sbjct: 701 VIIGNC*AIIVSTFFLWFRNILGYAYSN 784
>BQ124842 weakly similar to GP|19310633|gb unknown protein {Arabidopsis
thaliana}, partial (31%)
Length = 706
Score = 184 bits (468), Expect = 4e-47
Identities = 91/209 (43%), Positives = 138/209 (65%), Gaps = 2/209 (0%)
Frame = +2
Query: 178 MFC--LEWWSFEILTIVAGLLPNSQLETSVLSVCLSTTTLHYFIPHAIGASASTRVSNEL 235
+FC L+ W+FE++ +++GLLPN +ETSVLS+CL+T L + IP + S RVSNEL
Sbjct: 80 IFCDSLKVWTFELMVLMSGLLPNPVIETSVLSICLNTFGLAWMIPFGCSCAVSIRVSNEL 259
Query: 236 GAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVDYVTDMVPFLCV 295
G GNP A AVRVA+ I A+ + + R + G+ YS+DK+V+ YV+ M+P L +
Sbjct: 260 GGGNPNGASLAVRVALSISFIAALFMVLSMILARKVWGHLYSDDKQVIRYVSAMMPILAI 439
Query: 296 SVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLWMGTLT 355
S D+I LSG+ G G+Q IGAYVNLG++Y+VG P A L F + ++A GLW+G ++
Sbjct: 440 SSFLDAIQSTLSGVLAGCGWQKIGAYVNLGSFYVVGVPCAVVLAFFVHMHAMGLWLGIIS 619
Query: 356 GSILNVIILAVVTMLTDWQKEATKARERI 384
I+ + + T+ ++W++EA KA+ R+
Sbjct: 620 AFIVQTSLYIIFTIRSNWEEEAKKAQSRV 706
>AW773745 weakly similar to GP|19310633|gb unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 614
Score = 179 bits (453), Expect = 2e-45
Identities = 86/189 (45%), Positives = 126/189 (66%), Gaps = 2/189 (1%)
Frame = +3
Query: 1 MAGALETLCGQTYGAEEFSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIA 60
MA AL+TLCGQ+YGA+++ +G ++ AM L++V P++++W +L+ GQD EI+
Sbjct: 48 MASALDTLCGQSYGAKQYRMLGIHMQRAMFILMIVAIPLAIIWANTRSILIFLGQDHEIS 227
Query: 61 QAAREYCICLIPALFGHAVLQSLIRYFQIQSMIFPMVFSSIVILCLHVPICWCLVFKFGL 120
A Y ++P+LF + +LQ L R+ Q Q+++FPM+ SS V LH+P+CW +V+K G
Sbjct: 228 MEAGNYAKLMVPSLFAYGLLQCLNRFLQTQNIVFPMMLSSAVTTLLHLPLCWYMVYKSGF 407
Query: 121 GHVGAAFAIGIAYWLNVIWLGIYMKYSPACEKTKIVFSYNSLL--YIAEFCQFAIPSGLM 178
G GAA A I+YW+NVI L +Y+K+SP+C+KT FS +L I F + AIPS M
Sbjct: 408 GSGGAAIASSISYWVNVIILSLYVKFSPSCQKTWNGFSREALAPNNIPIFLKLAIPSAAM 587
Query: 179 FCLEWWSFE 187
CLE WSFE
Sbjct: 588 VCLEIWSFE 614
>AW980583 weakly similar to GP|16797803|db hypothetical membrane protein-1
{Marchantia polymorpha}, partial (21%)
Length = 631
Score = 164 bits (414), Expect = 7e-41
Identities = 81/204 (39%), Positives = 126/204 (61%)
Frame = +3
Query: 70 LIPALFGHAVLQSLIRYFQIQSMIFPMVFSSIVILCLHVPICWCLVFKFGLGHVGAAFAI 129
LIP + +L+ ++++ Q Q+++FP++ +S + LH CW + K G GAA A
Sbjct: 18 LIPNILASGILKCIVKFLQTQNIVFPLLVASGITSLLHCLNCWIWIVKLRHGIKGAAIAT 197
Query: 130 GIAYWLNVIWLGIYMKYSPACEKTKIVFSYNSLLYIAEFCQFAIPSGLMFCLEWWSFEIL 189
++ W + L Y+K+S +C+ T FS SL I +F + A PS +M CLE W +EI+
Sbjct: 198 CMSNWTYTVLLVFYIKFSSSCKSTWTGFSRESLHNIPQFLRIAFPSAIMVCLESWMYEIM 377
Query: 190 TIVAGLLPNSQLETSVLSVCLSTTTLHYFIPHAIGASASTRVSNELGAGNPRAAKGAVRV 249
+++G LPN +L+TSVL++C++ ++ + + +AS RVSNELGAGNPRAA AV V
Sbjct: 378 VLLSGTLPNPKLQTSVLAICMNIASVVWMLSSGFTGAASIRVSNELGAGNPRAAHLAVCV 557
Query: 250 AVIIGIAEAVIVSTLFLCFRNIIG 273
V++ I EA V T+ + RNI G
Sbjct: 558 VVVLNITEAFFVGTVMILLRNIWG 629
>TC80665 weakly similar to GP|19310633|gb|AAL85047.1 unknown protein
{Arabidopsis thaliana}, partial (31%)
Length = 894
Score = 162 bits (411), Expect = 2e-40
Identities = 81/152 (53%), Positives = 105/152 (68%)
Frame = +3
Query: 233 NELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVDYVTDMVPF 292
NELGAGNPRAA+ AV V V+I I E+++V + + RNI G AYSN++EVV YV M+P
Sbjct: 3 NELGAGNPRAARLAVYVVVVIAIIESIVVGAVIILIRNIWGYAYSNEEEVVKYVAIMLPI 182
Query: 293 LCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLWMG 352
+ VS D I LSG ARG G+Q IGAYVNLG+YYLVG P A L F L + KGLW+G
Sbjct: 183 IAVSNFLDGIQSVLSGTARGVGWQKIGAYVNLGSYYLVGIPAAVVLAFVLHVGGKGLWLG 362
Query: 353 TLTGSILNVIILAVVTMLTDWQKEATKARERI 384
+ + V+ L ++T+ TDW+KEA KA +R+
Sbjct: 363 IICALFVQVVSLTIITIRTDWEKEAKKATDRV 458
>TC78456 similar to GP|11994404|dbj|BAB02363.
gb|AAC28507.1~gene_id:MIL23.25~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (40%)
Length = 920
Score = 155 bits (391), Expect = 3e-38
Identities = 78/199 (39%), Positives = 121/199 (60%)
Frame = +1
Query: 186 FEILTIVAGLLPNSQLETSVLSVCLSTTTLHYFIPHAIGASASTRVSNELGAGNPRAAKG 245
F+IL ++AGLLP+ +L LS+C + + + I A+AS RVSNELGAGN ++A
Sbjct: 4 FQILVLLAGLLPHPELALDSLSICTTVSGWTFMISVGFQAAASVRVSNELGAGNSKSASF 183
Query: 246 AVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVDYVTDMVPFLCVSVSADSIICA 305
+V V +I I++ + L R++I ++ +EV V+++ P L +++ + +
Sbjct: 184 SVVVVTVISFIICAIIALVVLALRDVISYVFTEGEEVAAAVSNLSPLLALAIVLNGVQPV 363
Query: 306 LSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLWMGTLTGSILNVIILA 365
LSG+A G G+QT AYVN+G YY +G P+ LGF K AKG+W+G L G++L IIL
Sbjct: 364 LSGVAVGCGWQTFVAYVNVGCYYGLGIPLGAVLGFYFKFGAKGIWLGMLGGTVLQTIILM 543
Query: 366 VVTMLTDWQKEATKARERI 384
VT TDW E ++ +R+
Sbjct: 544 WVTFRTDWNNEVVESNKRL 600
>TC83102 similar to GP|11994404|dbj|BAB02363.
gb|AAC28507.1~gene_id:MIL23.25~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (36%)
Length = 1048
Score = 136 bits (343), Expect = 1e-32
Identities = 85/258 (32%), Positives = 133/258 (50%), Gaps = 4/258 (1%)
Frame = +3
Query: 131 IAYWLNVIWLGIYMKYSPACEKTKIVFSYNSLLYIAEFCQFAIPSGLMFCLEW-WSFEIL 189
IA W N++W +F+ N Y+ C F M LE+ ++ L
Sbjct: 15 IALWSNLLWP---------------LFTVNY**YVCNLCFFF--GEFM*SLEYIYNIGFL 143
Query: 190 TIVAGLLPNSQLETSVLSVCLSTTTLH---YFIPHAIGASASTRVSNELGAGNPRAAKGA 246
+ ++ +++C +TT+ + I A+AS RVSNELGA NP++A +
Sbjct: 144 *YIRYTPYTNKFSFLFITICDCSTTVSGWVFMISVGFNAAASVRVSNELGARNPKSASFS 323
Query: 247 VRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVDYVTDMVPFLCVSVSADSIICAL 306
V+V +I +VI + + L R++I ++ + V V+D+ P L +S+ + I L
Sbjct: 324 VKVVTVISFIISVIAALIVLALRDVISYVFTEGEVVAAAVSDLCPLLSLSLVLNGIQPVL 503
Query: 307 SGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLWMGTLTGSILNVIILAV 366
SG+A G G+Q AYVN+G YY+ G P+ LGF AKG+W+G L G+ + IIL
Sbjct: 504 SGVAVGCGWQAFVAYVNVGCYYIAGIPLGAGLGFYFNFGAKGIWLGMLGGTTMQTIILMW 683
Query: 367 VTMLTDWQKEATKARERI 384
VT TDW KE +A +R+
Sbjct: 684 VTFRTDWNKEVKEAAKRL 737
>TC82524 similar to GP|19310633|gb|AAL85047.1 unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 712
Score = 129 bits (323), Expect = 3e-30
Identities = 59/134 (44%), Positives = 90/134 (67%)
Frame = +2
Query: 1 MAGALETLCGQTYGAEEFSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIA 60
MA AL+T CGQ+YGA+++ +G ++ AM L++V P++++W +LL+ GQD EI+
Sbjct: 311 MASALDTFCGQSYGAKQYRMLGVHMQRAMFILMVVAIPLAVIWANTRSILLVLGQDPEIS 490
Query: 61 QAAREYCICLIPALFGHAVLQSLIRYFQIQSMIFPMVFSSIVILCLHVPICWCLVFKFGL 120
A Y ++P LF + +LQ L R+ Q Q+++FPM+FSS + LH+PICW +V+K GL
Sbjct: 491 IEAGSYAKLMVPCLFAYGLLQCLNRFLQTQNIVFPMMFSSAMTTLLHLPICWFMVYKSGL 670
Query: 121 GHVGAAFAIGIAYW 134
G AA A I+YW
Sbjct: 671 GSRXAAIANSISYW 712
>TC78455 similar to GP|11994404|dbj|BAB02363.
gb|AAC28507.1~gene_id:MIL23.25~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (48%)
Length = 1060
Score = 127 bits (319), Expect = 7e-30
Identities = 63/202 (31%), Positives = 118/202 (58%), Gaps = 1/202 (0%)
Frame = +1
Query: 1 MAGALETLCGQTYGAEEFSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIA 60
M A+ETLCGQ +GA+++ +G Y+ + + L + +++++IF + +L+ G+ +IA
Sbjct: 382 MGSAVETLCGQAFGAKKYEMLGIYLQRSTVLLTIAGLILTIIYIFSEPILIFLGESPKIA 561
Query: 61 QAAREYCICLIPALFGHAVLQSLIRYFQIQSMIFPMVFSSIVILCLHVPICWCLVFKFGL 120
AA + LIP +F +A+ + ++ Q QS++ P + S L +H+ + + ++++ GL
Sbjct: 562 SAASLFVFGLIPQIFAYAINFPIQKFLQAQSIVAPSAYISAATLVIHLVLSYVVIYQIGL 741
Query: 121 GHVGAAFAIGIAYWLNVIWLGIYMKYSPACEKTKIVFSYNSLLYIAEFCQFAIPSGLMFC 180
G +GA+ + I++W+ VI +Y+ S C+ T FS+ + + EF + + S +M C
Sbjct: 742 GLLGASLVLSISWWIIVIAQFVYIVKSEKCKHTWKGFSFQAFSGLPEFFKLSAASAVMLC 921
Query: 181 LEWWSFEILTI-VAGLLPNSQL 201
LE W L AGLLP+ +L
Sbjct: 922 LETWYLSNLXFCXAGLLPHPEL 987
>BG451836 weakly similar to PIR|A96736|A96 hypothetical protein F23N20.13
[imported] - Arabidopsis thaliana, partial (21%)
Length = 644
Score = 92.4 bits (228), Expect(2) = 2e-28
Identities = 42/84 (50%), Positives = 63/84 (75%)
Frame = +3
Query: 1 MAGALETLCGQTYGAEEFSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIA 60
M+ ALET CGQ YGA+++ K G + +A+I+LI+ C P+SL+W+ + KLL L GQD I+
Sbjct: 273 MSCALETQCGQAYGAKQYRKFGVQVYTAIISLIIACVPLSLLWLNLGKLLSLLGQDPLIS 452
Query: 61 QAAREYCICLIPALFGHAVLQSLI 84
Q A ++ +C+IPALF +A LQ+L+
Sbjct: 453 QEAGKFAMCMIPALFAYATLQALL 524
Score = 51.6 bits (122), Expect(2) = 2e-28
Identities = 23/39 (58%), Positives = 32/39 (81%)
Frame = +1
Query: 125 AAFAIGIAYWLNVIWLGIYMKYSPACEKTKIVFSYNSLL 163
AAF+IG +YWLNVI LG+YMK+S CEKT V+++N ++
Sbjct: 529 AAFSIGTSYWLNVIILGLYMKFSADCEKTP-VYNFNGVI 642
>BQ149337 weakly similar to GP|13272459|gb unknown protein {Arabidopsis
thaliana}, partial (21%)
Length = 622
Score = 117 bits (294), Expect = 6e-27
Identities = 58/160 (36%), Positives = 97/160 (60%)
Frame = +2
Query: 225 ASASTRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVD 284
A+AS RV+NELG G+ R AK ++ + + A I +FL + + +++D +V +
Sbjct: 8 AAASVRVANELGRGSSRDAKFSIVINALTSFAIGFIFFLIFLFLKKKLSYIFTSDSDVAN 187
Query: 285 YVTDMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKL 344
V D+ +L +S+ +S+ LSG++ G G+Q+I AYVN+G YYL+G P+ +G L
Sbjct: 188 AVGDLSFWLALSMLLNSVQPVLSGVSVGAGWQSIVAYVNIGCYYLIGIPVGVVIGVVYNL 367
Query: 345 NAKGLWMGTLTGSILNVIILAVVTMLTDWQKEATKARERI 384
KG+W+G L G+ + I+L ++T TDW K+ A+ R+
Sbjct: 368 GIKGIWIGMLFGTFVQTIMLIIITTKTDWDKQVEIAQWRV 487
>TC92785 weakly similar to GP|13937234|gb|AAK50109.1 AT5g38030/F16F17_30
{Arabidopsis thaliana}, partial (30%)
Length = 533
Score = 111 bits (277), Expect = 6e-25
Identities = 54/145 (37%), Positives = 87/145 (59%)
Frame = +1
Query: 240 PRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVDYVTDMVPFLCVSVSA 299
PR A+ ++ VAVI I ++++ + + R+ ++ DKEV D V D+ P L + V
Sbjct: 1 PRTARFSLVVAVITSILIGLLLALVLIISRDKYPAYFTTDKEVQDLVKDLTPLLALCVVI 180
Query: 300 DSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLWMGTLTGSIL 359
+++ LSG+A G G+Q AYVN+ YYL G P+ LG+ + L KG+W G ++G+IL
Sbjct: 181 NNVQPVLSGVAIGAGWQAAVAYVNIACYYLFGIPVGLILGYKVNLGVKGIWCGMMSGTIL 360
Query: 360 NVIILAVVTMLTDWQKEATKARERI 384
+L ++ T+W KEA+ A +RI
Sbjct: 361 QTCVLLIMVYKTNWNKEASLAEDRI 435
>BE319359 similar to GP|19310633|gb| unknown protein {Arabidopsis thaliana},
partial (20%)
Length = 313
Score = 109 bits (272), Expect = 2e-24
Identities = 54/103 (52%), Positives = 75/103 (72%), Gaps = 2/103 (1%)
Frame = +2
Query: 142 IYMKYSPACEKTKIVFSYNS--LLYIAEFCQFAIPSGLMFCLEWWSFEILTIVAGLLPNS 199
+Y+K+SP+C+KT FS + L I F + A+PS +M CLE WSFE++ +++GLLPN
Sbjct: 5 LYVKFSPSCKKTWTGFSKEAFALNNIPIFLKLAVPSAVMVCLEMWSFELMVLLSGLLPNP 184
Query: 200 QLETSVLSVCLSTTTLHYFIPHAIGASASTRVSNELGAGNPRA 242
+LETSVLS+ L+T+ L + IP + + S RVSNELGAGNPRA
Sbjct: 185 KLETSVLSISLNTSALVWMIPFGLSGAISIRVSNELGAGNPRA 313
>BF648997 weakly similar to GP|13384114|gb aberrant lateral root formation 5
{Arabidopsis thaliana}, partial (27%)
Length = 536
Score = 108 bits (271), Expect = 3e-24
Identities = 53/144 (36%), Positives = 87/144 (59%)
Frame = +3
Query: 1 MAGALETLCGQTYGAEEFSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIA 60
++GALETLCGQ +GA+E+ +G Y+ + I + IS++W + + +L+L Q +IA
Sbjct: 102 LSGALETLCGQGFGAKEYHMLGIYLQGSCIISFIFSIFISIIWFYTEHILVLLHQSQDIA 281
Query: 61 QAAREYCICLIPALFGHAVLQSLIRYFQIQSMIFPMVFSSIVILCLHVPICWCLVFKFGL 120
+ A Y LIP LF +++LQ+L+R+ Q QS++ P+V S + +HV I + V GL
Sbjct: 282 RTAALYMKFLIPGLFAYSILQNLLRFLQTQSVVMPLVILSAIPTLIHVGIAYGFVQWTGL 461
Query: 121 GHVGAAFAIGIAYWLNVIWLGIYM 144
+G A I W+++I L Y+
Sbjct: 462 NFIGGPVATSIXLWISMIMLXFYV 533
>BF648149 similar to PIR|T05135|T05 hypothetical protein F7H19.220 -
Arabidopsis thaliana, partial (42%)
Length = 642
Score = 106 bits (265), Expect = 1e-23
Identities = 70/211 (33%), Positives = 109/211 (51%), Gaps = 7/211 (3%)
Frame = +1
Query: 90 QSMIFPMVFSSIVILCLHVPICWCLVFKFGLGHVGAAFAIGIAYWLN---VIWLGIYMKY 146
QS+ P+ +S+ + + LH+PI + LV LG G A + W N V+ L IY+
Sbjct: 10 QSITLPLTYSATLSILLHIPINYFLVNVLQLGIRGIALG---SVWTNFNLVVSLIIYIWV 180
Query: 147 SPACEKTKIVFSYNSLLYIAEFCQFAIPSGLMFCLEWWSFEILTIVAGLLPNSQLETSVL 206
S +KT S AIPS + CLEWW +EI+ ++ GLL N + +
Sbjct: 181 SGTHKKTWSGISSACFKGWKSLLNLAIPSCISVCLEWWWYEIMILLCGLLLNPHATVASM 360
Query: 207 SVCLSTTTLHYFIPHAIGASASTRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIV--STL 264
V + TT L Y P ++ STRV NELGA NP+ AK +A I+G+ + ++ S L
Sbjct: 361 GVLIQTTALIYIFPSSLSFGVSTRVGNELGAENPQKAK----LAAIVGLCFSFVLGFSAL 528
Query: 265 FLCF--RNIIGNAYSNDKEVVDYVTDMVPFL 293
F F RNI +++D +++ + ++P +
Sbjct: 529 FFAFSVRNIWATMFTSDPQIIALTSMVLPII 621
>AW684439 weakly similar to PIR|F86285|F86 hypothetical protein AAD39644.1
[imported] - Arabidopsis thaliana, partial (32%)
Length = 510
Score = 102 bits (253), Expect = 3e-22
Identities = 46/91 (50%), Positives = 68/91 (74%)
Frame = +3
Query: 1 MAGALETLCGQTYGAEEFSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIA 60
M+ ALET CGQ YGA+++ K G I +A+ +LI+ C P+SL+WIF+ +LL+L GQD I+
Sbjct: 237 MSCALETQCGQAYGAKQYKKFGVQIYTAVFSLIIACLPLSLLWIFLGRLLILLGQDPLIS 416
Query: 61 QAAREYCICLIPALFGHAVLQSLIRYFQIQS 91
Q A ++ +C+IPALF +A L +L+ YF +QS
Sbjct: 417 QEAGKFSMCMIPALFAYATLXALVXYFLMQS 509
>TC80556 weakly similar to PIR|F86285|F86285 hypothetical protein AAD39644.1
[imported] - Arabidopsis thaliana, partial (31%)
Length = 661
Score = 102 bits (253), Expect = 3e-22
Identities = 48/93 (51%), Positives = 67/93 (71%)
Frame = +1
Query: 1 MAGALETLCGQTYGAEEFSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIA 60
MA LET CGQ YGA+++ +IG +++ +LILVC P+S +WI I+ +L+ GQD IA
Sbjct: 382 MASGLETTCGQAYGAKQYQRIGIQTYTSIFSLILVCLPLSFIWINIESILVFTGQDPLIA 561
Query: 61 QAAREYCICLIPALFGHAVLQSLIRYFQIQSMI 93
A + I LIPALF +A+LQ L+RYFQIQS++
Sbjct: 562 HEAGRFTIWLIPALFAYAILQPLVRYFQIQSLL 660
>BQ750943 weakly similar to GP|21627817|emb hypothetical protein {Aspergillus
fumigatus}, partial (7%)
Length = 711
Score = 99.8 bits (247), Expect = 2e-21
Identities = 60/167 (35%), Positives = 81/167 (47%)
Frame = -1
Query: 217 YFIPHAIGASASTRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAY 276
Y IP + +ASTR++N +GAG AAK +VA + V LF R + +
Sbjct: 711 YQIPFPVSIAASTRIANLIGAGLVDAAKTTGKVAFGAALTVGVFNVILFGGLRFHLPRLF 532
Query: 277 SNDKEVVDYVTDMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAY 336
SND+EV+ V +++P V D + G+ RG G +IG Y NL YY V P+++
Sbjct: 531 SNDEEVIAIVANVLPLCAVMQVFDGLAAGAHGLLRGIGRPSIGGYANLAVYYGVALPLSF 352
Query: 337 FLGFGLKLNAKGLWMGTLTGSILNVIILAVVTMLTDWQKEATKARER 383
F L KGLW G G L I LTDW K A +A R
Sbjct: 351 STAFLLDWKLKGLWFGVTVGLALVSAIEYAFVALTDWHKAAKEAETR 211
>TC93253 weakly similar to GP|14030731|gb|AAK53040.1 AT5g65380/MNA5_11
{Arabidopsis thaliana}, partial (25%)
Length = 633
Score = 85.9 bits (211), Expect = 2e-17
Identities = 42/132 (31%), Positives = 71/132 (52%), Gaps = 1/132 (0%)
Frame = +2
Query: 266 LCFRNIIGNAYSNDKEVVDYVTDMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLG 325
+ F ++ V++ V DM L V++ +S+ LSG+A G G+Q AYVN+G
Sbjct: 2 MIFHRQFAYIFTTSPPVLEAVNDMSILLAVTILLNSVQPILSGVAVGSGWQVFVAYVNIG 181
Query: 326 AYYLVGAPIAYFLGFGLKLNAKGLWMGTL-TGSILNVIILAVVTMLTDWQKEATKARERI 384
YYL+G P+ +G+ +G+W G + G+ + +IL +VT DW+ EA KAR +
Sbjct: 182 CYYLIGLPLGILMGWVFNTGVEGIWGGMIFGGTAIQTLILIIVTARCDWENEAKKARSSV 361
Query: 385 AEKPIEAHDGSI 396
++ + D +
Sbjct: 362 SKWSVTKPDDQL 397
>BG646347 similar to PIR|T05135|T05 hypothetical protein F7H19.220 -
Arabidopsis thaliana, partial (33%)
Length = 752
Score = 84.3 bits (207), Expect = 7e-17
Identities = 42/122 (34%), Positives = 70/122 (56%)
Frame = +3
Query: 1 MAGALETLCGQTYGAEEFSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIA 60
+A +E +CGQ +GA++F+ +G + ++ L+LV PIS W+++ K+LL F QD EIA
Sbjct: 384 LAVGMEPICGQAFGAKKFTLLGLCLQRTILLLLLVSIPISFSWLYMKKMLLFFNQDEEIA 563
Query: 61 QAAREYCICLIPALFGHAVLQSLIRYFQIQSMIFPMVFSSIVILCLHVPICWCLVFKFGL 120
A+ Y + IP L + + L Y + QS+ P+ + + LH+PI + L L
Sbjct: 564 TMAQTYILYSIPDLIAQSFIHPLRIYLRTQSITLPLTLCATFSILLHIPINYFLGIISYL 743
Query: 121 GH 122
G+
Sbjct: 744 GY 749
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.329 0.142 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,088,357
Number of Sequences: 36976
Number of extensions: 233982
Number of successful extensions: 1818
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 1794
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1815
length of query: 396
length of database: 9,014,727
effective HSP length: 98
effective length of query: 298
effective length of database: 5,391,079
effective search space: 1606541542
effective search space used: 1606541542
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144375.8


