

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
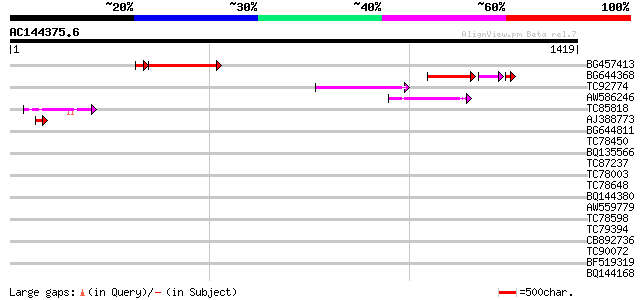
Query= AC144375.6 - phase: 0
(1419 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG457413 weakly similar to GP|13122061|em unnamed protein produc... 367 e-115
BG644368 similar to GP|12597768|g DNA ligase I putative {Arabid... 165 2e-50
TC92774 weakly similar to GP|6579215|gb|AAF18258.1| T23G18.1 {Ar... 149 1e-35
AW586246 weakly similar to GP|6579215|gb| T23G18.1 {Arabidopsis ... 139 6e-33
TC85818 homologue to GP|21261784|emb|CAD32500. protein kinase Ck... 72 1e-12
AJ388773 homologue to GP|20198018|gb| putative eukaryotic transl... 47 4e-05
BG644811 similar to GP|15866773|gb| origin recognition complex s... 36 0.093
TC78450 nodule-specific glycine-rich protein 3 [Medicago truncat... 35 0.16
BQ135566 35 0.27
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 33 1.0
TC78003 similar to GP|20146093|dbj|BAB88935. glucosyltransferase... 33 1.0
TC78648 similar to PIR|S51529|S51529 SPF1 protein - sweet potato... 32 1.3
BQ144380 similar to GP|1185397|gb|A SH3 domain binding protein {... 32 1.8
AW559779 32 1.8
TC78598 similar to SP|Q43207|FKB7_WHEAT 70 kDa peptidylprolyl is... 32 1.8
TC79394 weakly similar to PIR|T49019|T49019 probable RNA binding... 32 2.3
CB892736 homologue to PIR|D84745|D847 plastid protein [imported]... 31 3.0
TC90072 similar to PIR|T10237|T10237 RNA helicase RH16 - Arabido... 31 3.0
BF519319 homologue to GP|21553952|gb| unknown {Arabidopsis thali... 31 3.0
BQ144168 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 31 3.9
>BG457413 weakly similar to GP|13122061|em unnamed protein product {Zea
mays}, partial (7%)
Length = 646
Score = 367 bits (943), Expect(2) = e-115
Identities = 180/182 (98%), Positives = 180/182 (98%)
Frame = +3
Query: 348 REYVRFLKPKKVVPTVGLDVEKSDSKHVDKMRKYFAGLVDETANKHEFLKGFKQCDSGRS 407
REYVRFLKPKKVVPTVGLDVEKSDSKHVDKMRKYFAGLVDETANKHEFLKGFKQCDSGRS
Sbjct: 99 REYVRFLKPKKVVPTVGLDVEKSDSKHVDKMRKYFAGLVDETANKHEFLKGFKQCDSGRS 278
Query: 408 GFEVGKDVGNDTEPGHSVEKEVKPSDVGGDKSIDQDVAMSLSSCMGETCIEDPTLLNDEE 467
GFEVGKDVGNDTEPGHSVEKEVKPSDVGGDKSIDQDVAMSLSSCMGETCIEDPTLLNDEE
Sbjct: 279 GFEVGKDVGNDTEPGHSVEKEVKPSDVGGDKSIDQDVAMSLSSCMGETCIEDPTLLNDEE 458
Query: 468 KEKVVQELSCCLPTWVTRSQMLDLISISGSNVVEAVSNFFERETEFHEQVNSSQTPVPTH 527
KEKVVQELSCCLPTWVTRSQMLDLISISGSNVVEAVSNFFERETEFHEQVNS QTP PTH
Sbjct: 459 KEKVVQELSCCLPTWVTRSQMLDLISISGSNVVEAVSNFFERETEFHEQVNSXQTPXPTH 638
Query: 528 RS 529
RS
Sbjct: 639 RS 644
Score = 66.6 bits (161), Expect(2) = e-115
Identities = 29/31 (93%), Positives = 31/31 (99%)
Frame = +2
Query: 316 YEVKRDKFKVREKDSCKIHLVPYSEHSNYEE 346
++VKRDKFKVREKDSCKIHLVPYSEHSNYEE
Sbjct: 2 HQVKRDKFKVREKDSCKIHLVPYSEHSNYEE 94
>BG644368 similar to GP|12597768|g DNA ligase I putative {Arabidopsis
thaliana}, partial (12%)
Length = 641
Score = 165 bits (417), Expect(3) = 2e-50
Identities = 83/119 (69%), Positives = 98/119 (81%), Gaps = 1/119 (0%)
Frame = +1
Query: 1047 FTCEYKYDGQRAQIHKLVDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEV 1106
+T KYDGQRAQIHKL DGSV VFSRNGDE+TSRFPDLV++I ESC +TFI+DAEV
Sbjct: 7 YTVNTKYDGQRAQIHKLSDGSVRVFSRNGDETTSRFPDLVNIITESCDSRGATFIVDAEV 186
Query: 1107 VGIDRKNGCRIMSFQELSSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHLAD-PLK 1164
V IDR+NG ++MSFQELSSR RG KD+++ + IKV ICIFVFDIMFANGE L + PL+
Sbjct: 187 VAIDRQNGPKLMSFQELSSRERGSKDSIIALDKIKVEICIFVFDIMFANGEQLLNLPLR 363
Score = 40.4 bits (93), Expect(3) = 2e-50
Identities = 26/61 (42%), Positives = 36/61 (58%)
Frame = +3
Query: 1174 SPPKTKVSEGFVL**TTWVF*ICKRNIYRSR*CLLNM*SHIN*NKCFP*RCTALFM*RNY 1233
+P K K+ EGF+ * + +F + RN R *CL *SH *++ * CT FM*R++
Sbjct: 357 APSKAKIFEGFIW*RQSRLFRVRDRNDCRV**CLCR**SHTG*DEQLS**CTTCFM*RDH 536
Query: 1234 G 1234
G
Sbjct: 537 G 539
Score = 34.7 bits (78), Expect(3) = 2e-50
Identities = 17/25 (68%), Positives = 20/25 (80%)
Frame = +2
Query: 1242 WVFSIEAF*QMVKGQTRLCGRIK*Y 1266
WV +I+A * MVKGQ RLC RIK*+
Sbjct: 563 WVHTIKAL*CMVKGQERLC*RIK*F 637
>TC92774 weakly similar to GP|6579215|gb|AAF18258.1| T23G18.1 {Arabidopsis
thaliana}, partial (28%)
Length = 850
Score = 149 bits (375), Expect = 1e-35
Identities = 83/240 (34%), Positives = 131/240 (54%), Gaps = 3/240 (1%)
Frame = +1
Query: 765 YNPKEHACWRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVLPAVY 824
++P CW +P P+L L F ++ +E G+I T I+CN+ R+++ +PED++P VY
Sbjct: 148 FDPSSVVCWEKDKPVPFLFLCLAFDMINEESGRIVITDIVCNLLRTVIHATPEDLVPVVY 327
Query: 825 LCTNKIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQTQRL 884
L N+IA HE +EL IG + + AL EACG +IK Y + GDLG VA+ R +Q +
Sbjct: 328 LSANRIAPAHEGLELGIGDASIIKALAEACGRTEQQIKIQYKEKGDLGLVAKASRSSQSM 507
Query: 885 LAPPTPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRNLRI 944
+ P L I+ ++ I+ ++G S +KK I L+ + + E +++R L LRI
Sbjct: 508 MRKPDALTIRKVFKTFHLIAKESGKDSQEKKKNHIKSLLVAATDCEPLYIIRLLQTKLRI 687
Query: 945 GAMLRTVLPALAHAVVM---NSRPTVYEEGTAENLKAALQVLSVAVVEAYNILPNLDIIV 1001
G +T+L AL A V +S+P + E L +V Y++LP+ D IV
Sbjct: 688 GYAEQTLLTALGQAAVYTEEHSKPPPEIQSPFEELLNC*KV--------YSVLPDYDKIV 843
>AW586246 weakly similar to GP|6579215|gb| T23G18.1 {Arabidopsis thaliana},
partial (22%)
Length = 614
Score = 139 bits (351), Expect = 6e-33
Identities = 76/207 (36%), Positives = 121/207 (57%)
Frame = +3
Query: 949 RTVLPALAHAVVMNSRPTVYEEGTAENLKAALQVLSVAVVEAYNILPNLDIIVPTLMNKG 1008
+T+L AL A V + G L+ A ++ V +AY++LP+ D I+ ++ G
Sbjct: 21 QTLLSALGQAAVYTEEHSKPPPGIQSPLEEATKI----VKQAYSVLPDYDKIISAMLTDG 188
Query: 1009 IEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQRAQIHKLVDGSV 1068
+ + + PGIP++PML++ + +AL +N CEYKYDG+RAQIH +GSV
Sbjct: 189 VWLLAKTCNFTPGIPVEPMLSEKITSVSEALNKLRNAELICEYKYDGERAQIHYKENGSV 368
Query: 1069 LVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRKNGCRIMSFQELSSRGR 1128
++SR+ + +T +FPD+V + K S+F++D E+V DRK RI+ Q LS+R R
Sbjct: 369 EIYSRSSERNTGKFPDVVAAVSRLKKKNVSSFVLDCEIVAYDRKKQ-RILPMQVLSTRAR 545
Query: 1129 GGKDTLVTKESIKVGICIFVFDIMFAN 1155
KD V+ +IKV +C+F FD++ N
Sbjct: 546 --KDVAVS--NIKVDVCVFPFDLLHLN 614
>TC85818 homologue to GP|21261784|emb|CAD32500. protein kinase Ck2
regulatory subunit 2 {Nicotiana tabacum}, partial (44%)
Length = 827
Score = 72.4 bits (176), Expect = 1e-12
Identities = 60/201 (29%), Positives = 91/201 (44%), Gaps = 19/201 (9%)
Frame = +2
Query: 36 PPLPSSTIPHSKLIPNTRFLIDSF--RHTTPSSFTYFLSHFHSDHYSPLSSSWSHGIIFC 93
PP PS T +S + F DS RH +P +++ Y L+ S++HG I+C
Sbjct: 128 PPSPSITAAYSSI-----FCFDSSV*RHLSPQQTLIVVTY-----YQGLTKSFNHGKIYC 277
Query: 94 SPITSHLLINILHIPSPFVHPLSLNQSVVIDGSVVTLIDANHCPGAVQFLFK-------V 146
S +T+ L+ + IP +H L LNQ + I G VT +DANHCPG++ LF+ +
Sbjct: 278 SSVTARLVNMNIGIPYDKLHILPLNQKIEIAGISVTCLDANHCPGSILILFEPPNGKRSL 457
Query: 147 NETESPRY-------VHTGDFRFNREMLLDLNLGEF---IGADAVFLDTTYCHPKFVFPT 196
+T R H G F +L EF + D + D C P
Sbjct: 458 KQTVKSRM*VVLMERTHLGSHGF-----CNLRGNEFFCEVDDDYIQDDFNLCGLSSQVPY 622
Query: 197 QNESVDYIVDVVKECDGENVL 217
+ ++D I+DV + +N L
Sbjct: 623 YDYALDLILDVESSHEEQNEL 685
>AJ388773 homologue to GP|20198018|gb| putative eukaryotic translation
initiation factor 2 alpha subunit eIF2 {Arabidopsis
thaliana}, partial (19%)
Length = 609
Score = 47.4 bits (111), Expect = 4e-05
Identities = 19/29 (65%), Positives = 22/29 (75%)
Frame = -1
Query: 66 SFTYFLSHFHSDHYSPLSSSWSHGIIFCS 94
S YFL+HFHSDH LSS WSHG +FC+
Sbjct: 93 SEAYFLTHFHSDHTQGLSSXWSHGPLFCT 7
>BG644811 similar to GP|15866773|gb| origin recognition complex subunit 1
{Zea mays}, partial (4%)
Length = 784
Score = 36.2 bits (82), Expect = 0.093
Identities = 39/157 (24%), Positives = 67/157 (41%), Gaps = 18/157 (11%)
Frame = +3
Query: 524 VPTHRSCSSNDTSPLSKSNLKSFSSNDASPFSKSNL--NNTNSTTKKLDLFRSQESKLTN 581
+PT RS N SP++ + + ++P + N+ + +LD R +++ N
Sbjct: 117 LPTRRSTRLNSDSPITPNVIAESQIQQSTPRRGRRIGENSVPAAKSRLDFTRKEKAVTHN 296
Query: 582 LRKALSNQISPS----KRKKGSES-------KSNKKVKVKAKSESSGSKQATITKFFGK- 629
+ N+I + KR + +ES KS K K + G K + K GK
Sbjct: 297 SSIKVENEIDSAQFLRKRSRKAESDEIVFAPKSPDSSKSVKKRKKEGEKTVELKKRNGKG 476
Query: 630 -AMPVMPGDTQSDQFGSKPGE---SPEVEELVPTDAG 662
++ V T DQ +K + +VE++V T AG
Sbjct: 477 DSVKVSFAPTSPDQSETKKRKRKNEVKVEKMVVTRAG 587
>TC78450 nodule-specific glycine-rich protein 3 [Medicago truncatula]
Length = 941
Score = 35.4 bits (80), Expect = 0.16
Identities = 20/51 (39%), Positives = 27/51 (52%)
Frame = -3
Query: 30 PPTLPLPPLPSSTIPHSKLIPNTRFLIDSFRHTTPSSFTYFLSHFHSDHYS 80
PP L P P + P S+ + F++DSF TP+ + L HFH HYS
Sbjct: 324 PPPLFSSPYPQNDPPQSRQLSIPIFVLDSF--VTPTFLLHLLCHFH--HYS 184
>BQ135566
Length = 930
Score = 34.7 bits (78), Expect = 0.27
Identities = 34/101 (33%), Positives = 42/101 (40%), Gaps = 10/101 (9%)
Frame = +2
Query: 28 LPPPTLPL---------PPLPSSTIPHSKLIPNTRFLIDSFRHTTPSSFTYFLSHFHSDH 78
+P P +PL PP P S +PHS F PS F L +H
Sbjct: 320 IPKPHIPLTHHSHPNTTPPTPISPLPHS-----------LFPFCYPSPFQPHLLPYHIPF 466
Query: 79 YSPLSSSWSHGIIFCSPITSHLLINILHIPSPF-VHPLSLN 118
Y PLS S SH SPI+ +HI SP+ HP L+
Sbjct: 467 YHPLSHS-SH--YTTSPIS-------IHIHSPYTAHPPQLS 559
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 32.7 bits (73), Expect = 1.0
Identities = 45/239 (18%), Positives = 92/239 (37%)
Frame = +1
Query: 368 EKSDSKHVDKMRKYFAGLVDETANKHEFLKGFKQCDSGRSGFEVGKDVGNDTEPGHSVEK 427
E + K +K+++ DE ++ E + K + + E+ K+ G + E G E+
Sbjct: 166 ENEEIKDGEKIQQENEENKDEEKSQQENEEN-KDEEKSQQENELKKNEGGEKETGEITEE 342
Query: 428 EVKPSDVGGDKSIDQDVAMSLSSCMGETCIEDPTLLNDEEKEKVVQELSCCLPTWVTRSQ 487
+ K + ++ +D S+ G E ++++ ++ +E +
Sbjct: 343 KSKQENEETSETNSKDKENEESNQNGSDAKEQVGENHEQDSKQGTEETNGTEGGEKEEHD 522
Query: 488 MLDLISISGSNVVEAVSNFFERETEFHEQVNSSQTPVPTHRSCSSNDTSPLSKSNLKSFS 547
+ + S + V + N RE + N+S V SSN T + F
Sbjct: 523 KIKEDTSSDNQVQDGEKNNEAREENYSGD-NASSAVVDNKSQESSNKTE-------EQFD 678
Query: 548 SNDASPFSKSNLNNTNSTTKKLDLFRSQESKLTNLRKALSNQISPSKRKKGSESKSNKK 606
+ + F + N+N TT+ D +Q S+ K + + + + SES K+
Sbjct: 679 KKEKNEFELESQKNSNETTESTDSTITQNSQGNESEKDQAQTENDTPKGSASESDEQKQ 855
>TC78003 similar to GP|20146093|dbj|BAB88935. glucosyltransferase NTGT2
{Nicotiana tabacum}, partial (50%)
Length = 1740
Score = 32.7 bits (73), Expect = 1.0
Identities = 25/85 (29%), Positives = 40/85 (46%), Gaps = 3/85 (3%)
Frame = -3
Query: 42 TIPHSKLIPNTRFLID---SFRHTTPSSFTYFLSHFHSDHYSPLSSSWSHGIIFCSPITS 98
T P KL+ RF + SF + +SF +F +S +SPL + S ++ SP+T
Sbjct: 1531 TTPSKKLL---RFFSEEPPSFTASLANSFHFFPLLLNSSPFSPLPITTSKHLLISSPLTI 1361
Query: 99 HLLINILHIPSPFVHPLSLNQSVVI 123
+P H S+N ++VI
Sbjct: 1360 PSSFTSSSTLTPIFHTSSINFALVI 1286
>TC78648 similar to PIR|S51529|S51529 SPF1 protein - sweet potato, partial
(18%)
Length = 990
Score = 32.3 bits (72), Expect = 1.3
Identities = 16/41 (39%), Positives = 23/41 (56%)
Frame = +1
Query: 27 NLPPPTLPLPPLPSSTIPHSKLIPNTRFLIDSFRHTTPSSF 67
++ PP+LPL PS +P S T FLI F ++P+ F
Sbjct: 244 SVQPPSLPLSSSPSFHLPFSSTFSPTDFLISPFFLSSPNVF 366
>BQ144380 similar to GP|1185397|gb|A SH3 domain binding protein {Rattus
norvegicus}, partial (12%)
Length = 1269
Score = 32.0 bits (71), Expect = 1.8
Identities = 34/117 (29%), Positives = 48/117 (40%), Gaps = 4/117 (3%)
Frame = -2
Query: 5 SKSQTLESLNSTQLYLNALQSLNLPPPTLPLPPLPSSTIPHSK--LIPNTRFLIDSFRHT 62
S+S L S L +L PPP+L LP + S P+S LIP T + H
Sbjct: 1184 SRSPPLSPPLSRSYLLPHSSTLLPPPPSLSLPIVSSPACPYSPRILIPRTPLYLSHPLHA 1005
Query: 63 TPSSFTYFLSHFHSDHYSPLSSSWSHGI-IFCSPITSHLLINILHI-PSPFVHPLSL 117
+ S+ P+S S H I +F S +T + + I P P P+ L
Sbjct: 1004 SRSTL-------------PISRSSPHSIYLFSSALTPSQIADSSDIYPLPAPRPVGL 873
>AW559779
Length = 504
Score = 32.0 bits (71), Expect = 1.8
Identities = 38/160 (23%), Positives = 66/160 (40%), Gaps = 7/160 (4%)
Frame = +2
Query: 439 SIDQDVAMSLSSCMGETCIEDPTLLNDEEKEKVVQELSCCLPTWVTRSQMLDLISISGSN 498
S+ QDV+ +SS + L E + +S L T +T + L+ I SN
Sbjct: 8 SLLQDVSQIISSNTSKIISNTNPL---ETNNTISSNISKPLTT-ITNTNPLETIKSISSN 175
Query: 499 VVEAVSNFFERETEFHEQVNSSQTPVPTHRSCSSNDTSPLSK-------SNLKSFSSNDA 551
++ ++ ++ TP+ T +S SSN +SPLS + SS+ +
Sbjct: 176 ILSPLTTI------------TNTTPLETIKSISSNISSPLSTIINTTPLETFNTISSHIS 319
Query: 552 SPFSKSNLNNTNSTTKKLDLFRSQESKLTNLRKALSNQIS 591
SP + N N + + S LT + +L N ++
Sbjct: 320 SPL--ITIKNANPLETIKSISSNISSPLTTITNSLVNPLN 433
>TC78598 similar to SP|Q43207|FKB7_WHEAT 70 kDa peptidylprolyl isomerase (EC
5.2.1.8) (Peptidylprolyl cis-trans isomerase) (PPiase),
partial (92%)
Length = 2028
Score = 32.0 bits (71), Expect = 1.8
Identities = 23/75 (30%), Positives = 33/75 (43%)
Frame = +2
Query: 764 KYNPKEHACWRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVLPAV 823
K E A R + Y +FS ++EK + KA I CN+ + L +D A
Sbjct: 1418 KAGKHERASKRYEKAIRYFEYDSSFS--DEEKQQAKALKISCNLNDAACKLKLKDYKQAE 1591
Query: 824 YLCTNKIAADHENVE 838
LCT + D NV+
Sbjct: 1592 KLCTKVLELDSRNVK 1636
>TC79394 weakly similar to PIR|T49019|T49019 probable RNA binding protein -
Arabidopsis thaliana, partial (74%)
Length = 1909
Score = 31.6 bits (70), Expect = 2.3
Identities = 17/51 (33%), Positives = 30/51 (58%), Gaps = 6/51 (11%)
Frame = -3
Query: 6 KSQTLESLNSTQLYLNALQSLNLP------PPTLPLPPLPSSTIPHSKLIP 50
++Q + ++ Q +L L++ +LP PP LPLP LP + H++L+P
Sbjct: 428 EAQEAQRVSFCQPHLLHLRAYSLPLLLPLLPPLLPLPLLPPPLLLHTQLLP 276
>CB892736 homologue to PIR|D84745|D847 plastid protein [imported] -
Arabidopsis thaliana, partial (28%)
Length = 885
Score = 31.2 bits (69), Expect = 3.0
Identities = 29/75 (38%), Positives = 37/75 (48%), Gaps = 14/75 (18%)
Frame = +1
Query: 28 LPPP----TLPLPPLPS-STIPHSKLIPNTRFLIDSFR---HTTPSSFTYF------LSH 73
LPPP TL + L S ST+P S L+P R L S R TPSS F ++
Sbjct: 10 LPPPYTTVTLSMAKLLSLSTLPSSLLLPPNRLLSSSTRRPSQVTPSSSFRFTTVQCRVNR 189
Query: 74 FHSDHYSPLSSSWSH 88
+ YSPL+S S+
Sbjct: 190AGNSTYSPLNSGNSN 234
>TC90072 similar to PIR|T10237|T10237 RNA helicase RH16 - Arabidopsis
thaliana, partial (28%)
Length = 814
Score = 31.2 bits (69), Expect = 3.0
Identities = 24/68 (35%), Positives = 34/68 (49%), Gaps = 3/68 (4%)
Frame = +1
Query: 789 SLLEDEKGKIKATSILCNMFRSLL---VLSPEDVLPAVYLCTNKIAADHENVELNIGGSL 845
SLLE K +KA + NM + L ++ P DVL + C K ++ +I GSL
Sbjct: 454 SLLELCKVPLKAVQLNSNMLATDLQAALVGPPDVLISTPACIAKCLSNSVLQAASINGSL 633
Query: 846 VTTALEEA 853
T L+EA
Sbjct: 634 ETLVLDEA 657
>BF519319 homologue to GP|21553952|gb| unknown {Arabidopsis thaliana},
partial (19%)
Length = 546
Score = 31.2 bits (69), Expect = 3.0
Identities = 28/112 (25%), Positives = 50/112 (44%), Gaps = 3/112 (2%)
Frame = +2
Query: 662 GNMYKQEIDQFMQI---INGDESLKKQAITIIEEAKGDINKALDIYYSNSCNLGEREISV 718
G++ ++EI+ F + I G +Q + +E+AK L + +
Sbjct: 35 GSLVRKEIEYFTKFELDIRGISQKPQQHLVDMEKAKKPSFWFLPTTWKKRKQRSSTNRFI 214
Query: 719 QGECKVDRPLEKKYVSKELNVIPDISMHRVLRDNVDATHVSLPSDKYNPKEH 770
+ C V E Y S + N IP S +++LRD+++ H+ SD K+H
Sbjct: 215 RKICSVT---EVDYESNQFNRIPGYS-YKILRDDINNFHL*AGSDLLQ*KKH 358
>BQ144168 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (9%)
Length = 1381
Score = 30.8 bits (68), Expect = 3.9
Identities = 13/20 (65%), Positives = 14/20 (70%)
Frame = +3
Query: 25 SLNLPPPTLPLPPLPSSTIP 44
S + PPP LPLPPLPS P
Sbjct: 3 SFSPPPPPLPLPPLPSHPPP 62
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.327 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,237,994
Number of Sequences: 36976
Number of extensions: 698196
Number of successful extensions: 6100
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 4475
Number of HSP's successfully gapped in prelim test: 276
Number of HSP's that attempted gapping in prelim test: 1316
Number of HSP's gapped (non-prelim): 5092
length of query: 1419
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1311
effective length of database: 5,021,319
effective search space: 6582949209
effective search space used: 6582949209
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC144375.6


